The ACCESS-NRI Evaluation Frameworks
Overview
Teaching: 20 min
Exercises: 30 min
Compatibility:Questions
What are the ACCESS-NRI supported evaluation frameworks?
How do I get started?
Where can I find help?
Objectives
The Model Evaluation and Diagnostics (MED) Team is Here to Help!
We support infrastructure (software + data) and provide technical support / training to the ACCESS community.
-
Tool Deployment on NCI Gadi: We make sure essential tools like ESMValTool and ILAMB are ready to go on NCI Gadi, so your research workflows run smoothly.
-
Evaluation Tools & Recipes: We develop, support, and fine-tune evaluation tools and scripts, ensuring they’re up to scratch.
-
Publication & Dissemination: Need to get your work out there? We assist with publishing and sharing your evaluation scripts.
-
Training: Whether you’re a newbie or a pro, we offer training for all levels to help you make the most of our tools and resources.
-
Community Hub: We’re your go-to for collaboration and knowledge-sharing, keeping the ACCESS ecosystem thriving.
If you need support, the MED team is here to help!
ACCESS-NRI Evaluation tools and infrastructure
Here is the current list of tools and supporting infrastructure under the ACCESS-NRI Model Evaluation and Diagnostics team responsibility:
- MED Conda Environments
- ESMValTool-Workflow
- ILAMB-Workflow
- ACCESS MED Diagnostics
- ACCESS-NRI Intake catalogue
- ACCESS-NRI Data Replicas for Model Evaluation and Diagnostics
MED Conda Environments
To ensure effective and efficient evaluation of model outputs, it is crucial to have a well-maintained and reliable analysis environment on the NCI Gadi supercomputer. Our approach involves releasing tools within containerized Conda environments, providing a consistent and dependable platform for users. These containerized environments simplify the deployment process, ensuring that all necessary dependencies and configurations are included, which minimizes setup time and potential issues.
ESMValTool-Workflow
ESMValTool-workflow is the ACCESS-NRI software and data infrastructure that enables the ESMValTool evaluation framework on NCI Gadi. It includes:
- The ESMValCore Python packages: This core library is designed to facilitate the preprocessing of climate data, offering a structured and efficient way to handle complex datasets.
- The ESMValTool collection of recipes, diagnostics and observation CMORisers.
- A data pool of CMORised observational datasets.
ESMValTool-workflow is configured to use the existing NCI supported CMIP data collections.
ESMValTool meets the community’s need for a robust, reliable, and reproducible framework to evaluate ACCESS climate models. Specifically developed with CMIP evaluation in mind, the software is well-suited for this purpose.
How do I get started?
The ESMValCore and ESMValTool python tools and their dependencies are deployed on Gadi within an ESMValTool-workflow containerized Conda environment that can be loaded as a module.
Using the command line and PBS jobs
If you have carefully completed the requirements, you should already be a member of the
xp65project and be ready to go.module use /g/data/xp65/public/modules # Load the ESMValTool-Workflow: module load esmvaltool-workflowUsing ARE
If you have carefully completed the requirements, you should already be a member of the
xp65project and be ready to go.
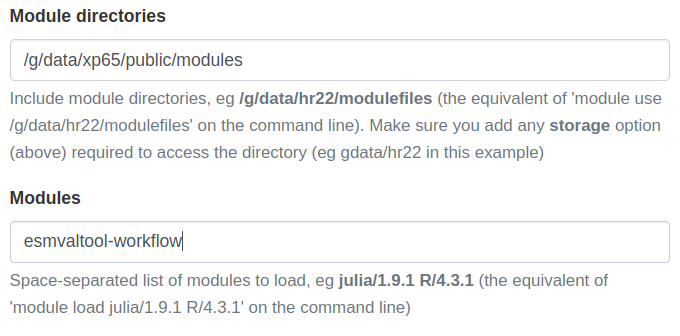
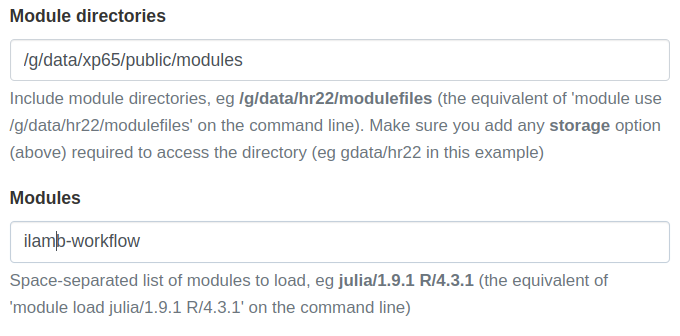
ILAMB-Workflow
The International Land Model Benchmarking (ILAMB) project is a model-data intercomparison and integration project designed to improve the performance of land models and, in parallel, improve the design of new measurement campaigns to reduce uncertainties associated with key land surface processes.
The ACCESS-NRI Model Evaluation and Diagnostics team is releasing and supporting NCI configuration of ILAMB under the name ILAMB-workflow.
ILAMB-workflow is the ACCESS-NRI software and data infrastructure that enables the ILAMB evaluation framework on NCI Gadi. It includes:
- the ILAMB Python packages
- a series of ILAMB outputs for ACCESS model evaluation and
- the ILAMB-Data collection of observational datasets.
ILAMB-workflow is configured to use the existing NCI supported CMIP data collections.
ILAMB addresses the needs of the Land community for a robust, reliable, and reproducible framework for evaluating land surface models.
How do I get started?
The ILAMB python tool and its dependencies are deployed on Gadi within an ILAMB-workflow containerized Conda environment that can be loaded as a module.
Using the command line and PBS jobs
If you have carefully completed the requirements, you should already be a member of the
xp65project and be ready to go.module use /g/data/xp65/public/modules # Load the ILAMB-Workflow: module load ilamb-workflowUsing ARE
If you have carefully completed the requirements, you should already be a member of the
xp65project and be ready to go.
Key Points
Introducing ESMValTool
Overview
Teaching: 5 min
Exercises: 10 min
Compatibility:Questions
What is ESMValTool?
Who are the people behind ESMValTool?
Objectives
Familiarize with ESMValTool
Synchronize expectations
What is ESMValTool?
This tutorial is a first introduction to ESMValTool. Before diving into the technical steps, let’s talk about what ESMValTool is all about.
What is ESMValTool?
What do you already know about or expect from ESMValTool?
ESMValTool is…
EMSValTool is many things, but in this tutorial we will focus on the following traits:
✓ A Python-based preprocessing framework
✓ A Standardised framework for climate data analysis
✓ A collection of diagnostics for reproducible climate science
✓ A community effort
A Python-based preprocessing framework
ESMValTool is powered by ESMValCore, a powerfull python-based workflow engine that facilitates CMIP analysis. ESMValCore implements the core functionality of ESMValTool: it takes care of finding, opening, checking, fixing, concatenating, and preprocessing CMIP data and several other supported datasets. ESMValCore has matured as a reliable foundation for the ESMValTool with recent addition making it attractive as a lightweight approach to CMIP evaluation.
A common scenario consist in visualising the global temperature of an historical run over a 2 year period. To do so, you need first to:
- Find the data
- Extract the period of interest
- Calculate the mean
- Convert the units to degrees celsius
- Finally Plot the data
The following example illustrate how to leverage ESMValCore, the engine powering the ESMValTool collection of recipes, to quickly load CMIP data and do some analysis on them.
from esmvalcore.dataset import Dataset
from esmvalcore.preprocessor import extract_time
from esmvalcore.preprocessor import climate_statistics
from esmvalcore.preprocessor import convert_units
dataset = Dataset(
short_name='tas',
project='CMIP6',
mip="Amon",
exp="historical",
ensemble="r1i1p1f1",
dataset='ACCESS-ESM1-5',
grid="gn"
)
temperature = dataset.load()
temperature_1990_1991 = extract_time(temperature, start_year=1990, start_month=1, start_day=1, end_year=1991, end_month=1, end_day=1)
temperature_weighted_mean = climate_statistics(temperature_1990_1991, operator="mean")
temperature_celsius = convert_units(temperature_weighted_mean, units="degrees_C")
Example Plots
ESMValCore uses Iris Cube to manipulate data. Iris can thus be used to quickly plot the data in a notebook, but you could use your package of choice.
import cartopy.crs as ccrs import matplotlib.pyplot as plt from matplotlib import colormaps import iris import iris.plot as iplt import iris.quickplot as qplt # Load a Cynthia Brewer palette. brewer_cmap = colormaps["brewer_OrRd_09"] # Create a figure plt.figure(figsize=(12, 5)) # Plot #1: countourf with axes longitude from -180 to 180 proj = ccrs.PlateCarree(central_longitude=0.0) plt.subplot(121, projection=proj) qplt.contourf(temperature_weighted_mean, brewer_cmap.N, cmap=brewer_cmap) plt.gca().coastlines() # Plot #2: contourf with axes longitude from 0 to 360 proj = ccrs.PlateCarree(central_longitude=-180.0) plt.subplot(122, projection=proj) qplt.contourf(temperature_weighted_mean, brewer_cmap.N, cmap=brewer_cmap) plt.gca().coastlines() iplt.show()
Exercises
ESMValCore has a growing collection of preprocessors, have a look at the documentation and see what is available.
- Open an ARE session and run the above example.
- See if you can load other datasets
- change the time period
- Add a new preprocessing step
A Standardised framework for climate data analysis
ESMValTool is a software project that was designed by and for climate scientists to evaluate CMIP data in a standardized and reproducible manner.
The central component of ESMValTool that we will see in this tutorial is the recipe. Any ESMValTool recipe is basically a set of instructions to reproduce a certain result. The basic structure of a recipe is as follows:
- Documentation with relevant (citation) information
- Datasets that should be analysed
- Preprocessor steps that must be applied
- Diagnostic scripts performing more specific evaluation steps
An example recipe could look like this:
documentation:
title: This is an example recipe.
description: Example recipe
authors:
- lastname_firstname
datasets:
- {dataset: ACCESS-CM2, project: CMIP6, exp: historical, mip: Amon,
ensemble: r1i1p1f1, start_year: 1960, end_year: 2005}
preprocessors:
global_mean:
area_statistics:
operator: mean
diagnostics:
average_plot:
description: plot of global mean temperature change
variables:
temperature:
short_name: tas
preprocessor: global_mean
scripts: examples/diagnostic.py
Understanding the different section of the recipe
Try to figure out the meaning of the different dataset keys. Hint: they can be found in the documentation of ESMValTool.
Solution
The keys are explained in the ESMValTool documentation, in the
Recipe section, under datasets
A collection of diagnostics for reproducible climate science
More than a tool, ESMValTool is a collection of publicly available recipes and diagnostic scripts. This makes it possible to easily reproduce important results.
Explore the available recipes
Go to the ESMValTool Documentation webpage and explore the
Available recipessection. Which recipe(s) would you like to try?
A community effort
ESMValTool is built and maintained by an active community of scientists and software engineers. It is an open source project to which anyone can contribute. Many of the interactions take place on GitHub. Here, we briefly introduce you to some of the most important pages.
Meet the ESMValGroup
Go to github.com/ESMValGroup. This is the GitHub page of our ‘organization’. Have a look around. How many collaborators are there? Do you know any of them?
Near the top of the page there are 2 pinned repositories: ESMValTool and ESMValCore. Visit each of the repositories. How many people have contributed to each of them? Can you also find out how many people have contributed to this tutorial?
Issues and pull requests
Go back to the repository pages of ESMValTool or ESMValCore. There are tabs for ‘issues’ and ‘pull requests’. You can use the labels to navigate them a bit more. How many open issues are about enhancements of ESMValTool? And how many bugs have been fixed in ESMValCore? There is also an ‘insights’ tab, where you can see a summary of recent activity. How many issues have been opened and closed in the past month?
Conclusion
This concludes the introduction of the tutorial. You now have a basic knowledge of ESMValTool and its community. The following episodes will walk you through the installation, configuration and running your first recipes.
Key Points
ESMValTool provides a reliable interface to analyse and evaluate climate data
A large collection of recipes and diagnostic scripts is already available
ESMValTool is built and maintained by an active community of scientists and developers
Running your first recipe
Overview
Teaching: 15 min
Exercises: 15 min
Compatibility:Questions
How to run a recipe?
What happens when I run a recipe?
Objectives
Run an existing ESMValTool recipe
Examine the log information
Navigate the output created by ESMValTool
Make small adjustments to an existing recipe
This episode describes how ESMValTool recipes work, how to run a recipe and how to explore the recipe output. By the end of this episode, you should be able to run your first recipe, look at the recipe output, and make small modifications.
Import module in GADI
You may want to open VS Code with a remote SSH connection to Gadi and use the VS Code terminal, then you can later view the recipe file. Refer to VS Code setup.
In a terminal with an SSH connection into Gadi, load the module to use ESMValTool on Gadi.
module use /g/data/xp65/public/modules
module load esmvaltool-workflow
Running an existing recipe
The recipe format has briefly been introduced in the Introduction episode. To see all the recipes that are shipped with ESMValTool, type
esmvaltool recipes list
We will start by running examples/recipe_python.yml. This is the command with ESMValTool installed.
esmvaltool run examples/recipe_python.yml
On Gadi, this can be done using the esmvaltool-workflow wrapper in the loaded module.
esmvaltool-workflow run examples/recipe_python.yml
or if you have the user configuration file in your current directory then
esmvaltool-workflow run --config_file ./config-user.yml examples/recipe_python.yml
You should see that Gadi has created a PBS job to run the recipe. You can check your
queue status with qstat.
[fc6164@gadi-login-01 fc6164]$ module load esmvaltool
Welcome to the ACCESS-NRI ESMValTool-Workflow
enter command `esmvaltool-workflow` for help
Loading esmvaltool/workflow_v1.2
Loading requirement: singularity conda/esmvaltool-0.4
[fc6164@gadi-login-01 fc6164]$ esmvaltool-workflow run recipe_python.yml
conda/esmvaltool-0.4
123732363.gadi-pbs
Running recipe: recipe_python.yml
[fc6164@gadi-login-01 fc6164]$ qstat
Job id Name User Time Use S Queue
--------------------- ---------------- ---------------- -------- - -----
123732363.gadi-pbs recipe_python fc6164 0 Q normal-exec
[fc6164@gadi-login-01 fc6164]$
If everything is okay, the final log message should be “Run was successful”. The exact output varies depending on your machine, this is an example of a successful log output below.
Example output
2024-05-15 07:04:08,041 UTC [134535] INFO ______________________________________________________________________ _____ ____ __ ____ __ _ _____ _ | ____/ ___|| \/ \ \ / /_ _| |_ _|__ ___ | | | _| \___ \| |\/| |\ \ / / _` | | | |/ _ \ / _ \| | | |___ ___) | | | | \ V / (_| | | | | (_) | (_) | | |_____|____/|_| |_| \_/ \__,_|_| |_|\___/ \___/|_| ______________________________________________________________________ ESMValTool - Earth System Model Evaluation Tool. http://www.esmvaltool.org CORE DEVELOPMENT TEAM AND CONTACTS: Birgit Hassler (Co-PI; DLR, Germany - birgit.hassler@dlr.de) Alistair Sellar (Co-PI; Met Office, UK - alistair.sellar@metoffice.gov.uk) Bouwe Andela (Netherlands eScience Center, The Netherlands - b.andela@esciencecenter.nl) Lee de Mora (PML, UK - ledm@pml.ac.uk) Niels Drost (Netherlands eScience Center, The Netherlands - n.drost@esciencecenter.nl) Veronika Eyring (DLR, Germany - veronika.eyring@dlr.de) Bettina Gier (UBremen, Germany - gier@uni-bremen.de) Remi Kazeroni (DLR, Germany - remi.kazeroni@dlr.de) Nikolay Koldunov (AWI, Germany - nikolay.koldunov@awi.de) Axel Lauer (DLR, Germany - axel.lauer@dlr.de) Saskia Loosveldt-Tomas (BSC, Spain - saskia.loosveldt@bsc.es) Ruth Lorenz (ETH Zurich, Switzerland - ruth.lorenz@env.ethz.ch) Benjamin Mueller (LMU, Germany - b.mueller@iggf.geo.uni-muenchen.de) Valeriu Predoi (URead, UK - valeriu.predoi@ncas.ac.uk) Mattia Righi (DLR, Germany - mattia.righi@dlr.de) Manuel Schlund (DLR, Germany - manuel.schlund@dlr.de) Breixo Solino Fernandez (DLR, Germany - breixo.solinofernandez@dlr.de) Javier Vegas-Regidor (BSC, Spain - javier.vegas@bsc.es) Klaus Zimmermann (SMHI, Sweden - klaus.zimmermann@smhi.se) For further help, please read the documentation at http://docs.esmvaltool.org. Have fun! 2024-05-15 07:04:08,044 UTC [134535] INFO Package versions 2024-05-15 07:04:08,044 UTC [134535] INFO ---------------- 2024-05-15 07:04:08,044 UTC [134535] INFO ESMValCore: 2.10.0 2024-05-15 07:04:08,044 UTC [134535] INFO ESMValTool: 2.10.0 2024-05-15 07:04:08,044 UTC [134535] INFO ---------------- 2024-05-15 07:04:08,044 UTC [134535] INFO Using config file /pfs/lustrep1/users/username/esmvaltool_tutorial/config-user.yml 2024-05-15 07:04:08,044 UTC [134535] INFO Writing program log files to: /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/main_log.txt /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/main_log_debug.txt 2024-05-15 07:04:08,503 UTC [134535] INFO Using default ESGF configuration, configuration file /users/username/.esmvaltool/esgf-pyclient.yml not present. 2024-05-15 07:04:08,504 UTC [134535] WARNING ESGF credentials missing, only data that is accessible without logging in will be available. See https://esgf.github.io/esgf-user-support/user_guide.html for instructions on how to create an account if you do not have one yet. Next, configure your system so esmvaltool can use your credentials. This can be done using the keyring package, or you can just enter them in /users/username/.esmvaltool/esgf-pyclient.yml. keyring ======= First install the keyring package (requires a supported backend, see https://pypi.org/project/keyring/): $ pip install keyring Next, set your username and password by running the commands: $ keyring set ESGF hostname $ keyring set ESGF username $ keyring set ESGF password To check that you entered your credentials correctly, run: $ keyring get ESGF hostname $ keyring get ESGF username $ keyring get ESGF password configuration file ================== You can store the hostname, username, and password or your OpenID account in a plain text in the file /users/username/.esmvaltool/esgf-pyclient.yml like this: logon: hostname: "your-hostname" username: "your-username" password: "your-password" or your can configure an interactive log in: logon: interactive: true Note that storing your password in plain text in the configuration file is less secure. On shared systems, make sure the permissions of the file are set so only you can read it, i.e. $ ls -l /users/username/.esmvaltool/esgf-pyclient.yml shows permissions -rw-------. 2024-05-15 07:04:09,067 UTC [134535] INFO Starting the Earth System Model Evaluation Tool at time: 2024-05-15 07:04:09 UTC 2024-05-15 07:04:09,068 UTC [134535] INFO ---------------------------------------------------------------------- 2024-05-15 07:04:09,068 UTC [134535] INFO RECIPE = /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/recipes/examples/recipe_python.yml 2024-05-15 07:04:09,068 UTC [134535] INFO RUNDIR = /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run 2024-05-15 07:04:09,069 UTC [134535] INFO WORKDIR = /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/work 2024-05-15 07:04:09,069 UTC [134535] INFO PREPROCDIR = /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/preproc 2024-05-15 07:04:09,069 UTC [134535] INFO PLOTDIR = /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/plots 2024-05-15 07:04:09,069 UTC [134535] INFO ---------------------------------------------------------------------- 2024-05-15 07:04:09,069 UTC [134535] INFO Running tasks using at most 256 processes 2024-05-15 07:04:09,069 UTC [134535] INFO If your system hangs during execution, it may not have enough memory for keeping this number of tasks in memory. 2024-05-15 07:04:09,070 UTC [134535] INFO If you experience memory problems, try reducing 'max_parallel_tasks' in your user configuration file. 2024-05-15 07:04:09,070 UTC [134535] WARNING Using the Dask basic scheduler. This may lead to slow computations and out-of-memory errors. Note that the basic scheduler may still be the best choice for preprocessor functions that are not lazy. In that case, you can safely ignore this warning. See https://docs.esmvaltool.org/projects/ESMValCore/en/latest/quickstart/configure.html#dask-distributed-configuration for more information. 2024-05-15 07:04:09,113 UTC [134535] WARNING 'default' rootpaths '/users/username/climate_data' set in config-user.yml do not exist 2024-05-15 07:04:10,648 UTC [134535] INFO Creating tasks from recipe 2024-05-15 07:04:10,648 UTC [134535] INFO Creating tasks for diagnostic map 2024-05-15 07:04:10,648 UTC [134535] INFO Creating diagnostic task map/script1 2024-05-15 07:04:10,649 UTC [134535] INFO Creating preprocessor task map/tas 2024-05-15 07:04:10,649 UTC [134535] INFO Creating preprocessor 'to_degrees_c' task for variable 'tas' 2024-05-15 07:04:11,066 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP6, BCC-ESM1, CMIP, historical, r1i1p1f1, gn, v20181214 2024-05-15 07:04:11,405 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP5, bcc-csm1-1, historical, r1i1p1, v1 2024-05-15 07:04:11,406 UTC [134535] INFO PreprocessingTask map/tas created. 2024-05-15 07:04:11,406 UTC [134535] INFO Creating tasks for diagnostic timeseries 2024-05-15 07:04:11,406 UTC [134535] INFO Creating diagnostic task timeseries/script1 2024-05-15 07:04:11,406 UTC [134535] INFO Creating preprocessor task timeseries/tas_amsterdam 2024-05-15 07:04:11,406 UTC [134535] INFO Creating preprocessor 'annual_mean_amsterdam' task for variable 'tas_amsterdam' 2024-05-15 07:04:11,428 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP6, BCC-ESM1, CMIP, historical, r1i1p1f1, gn, v20181214 2024-05-15 07:04:11,452 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP5, bcc-csm1-1, historical, r1i1p1, v1 2024-05-15 07:04:11,455 UTC [134535] INFO PreprocessingTask timeseries/tas_amsterdam created. 2024-05-15 07:04:11,455 UTC [134535] INFO Creating preprocessor task timeseries/tas_global 2024-05-15 07:04:11,455 UTC [134535] INFO Creating preprocessor 'annual_mean_global' task for variable 'tas_global' 2024-05-15 07:04:11,814 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP6, BCC-ESM1, CMIP, historical, r1i1p1f1, gn, v20181214, supplementaries: areacella, fx, 1pctCO2, v20190613 2024-05-15 07:04:12,184 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP5, bcc-csm1-1, historical, r1i1p1, v1, supplementaries: areacella, fx, r0i0p0 2024-05-15 07:04:12,186 UTC [134535] INFO PreprocessingTask timeseries/tas_global created. 2024-05-15 07:04:12,187 UTC [134535] INFO These tasks will be executed: timeseries/script1, timeseries/tas_global, map/script1, map/tas, timeseries/tas_amsterdam 2024-05-15 07:04:12,204 UTC [134535] INFO Wrote recipe with version numbers and wildcards to: file:///users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/recipe_python_filled.yml 2024-05-15 07:04:12,204 UTC [134535] INFO Will download 129.2 MB Will download the following files: 50.85 KB ESGFFile:CMIP6/CMIP/BCC/BCC-ESM1/1pctCO2/r1i1p1f1/fx/areacella/gn/v20190613/areacella_fx_BCC-ESM1_1pctCO2_r1i1p1f1_gn.nc on hosts ['aims3.llnl.gov', 'cmip.bcc.cma.cn', 'esgf-data04.diasjp.net', 'esgf.nci.org.au', 'esgf3.dkrz.de'] 64.95 MB ESGFFile:CMIP6/CMIP/BCC/BCC-ESM1/historical/r1i1p1f1/Amon/tas/gn/v20181214/tas_Amon_BCC-ESM1_historical_r1i1p1f1_gn_185001-201412.nc on hosts ['aims3.llnl.gov', 'cmip.bcc.cma.cn', 'esgf-data04.diasjp.net', 'esgf.ceda.ac.uk', 'esgf.nci.org.au', 'esgf3.dkrz.de'] 44.4 KB ESGFFile:cmip5/output1/BCC/bcc-csm1-1/historical/fx/atmos/fx/r0i0p0/v1/areacella_fx_bcc-csm1-1_historical_r0i0p0.nc on hosts ['aims3.llnl.gov', 'esgf.ceda.ac.uk', 'esgf2.dkrz.de'] 64.15 MB ESGFFile:cmip5/output1/BCC/bcc-csm1-1/historical/mon/atmos/Amon/r1i1p1/v1/tas_Amon_bcc-csm1-1_historical_r1i1p1_185001-201212.nc on hosts ['aims3.llnl.gov', 'esgf.ceda.ac.uk', 'esgf2.dkrz.de'] Downloading 129.2 MB.. 2024-05-15 07:04:14,074 UTC [134535] INFO Downloaded /users/username/climate_data/cmip5/output1/BCC/bcc-csm1-1/historical/fx/atmos/fx/r0i0p0/v1/areacella_fx_bcc-csm1-1_historical_r0i0p0.nc (44.4 KB) in 1.84 seconds (24.09 KB/s) from aims3.llnl.gov 2024-05-15 07:04:14,109 UTC [134535] INFO Downloaded /users/username/climate_data/CMIP6/CMIP/BCC/BCC-ESM1/1pctCO2/r1i1p1f1/fx/areacella/gn/v20190613/areacella_fx_BCC-ESM1_1pctCO2_r1i1p1f1_gn.nc (50.85 KB) in 1.88 seconds (27 KB/s) from aims3.llnl.gov 2024-05-15 07:04:20,505 UTC [134535] INFO Downloaded /users/username/climate_data/CMIP6/CMIP/BCC/BCC-ESM1/historical/r1i1p1f1/Amon/tas/gn/v20181214/tas_Amon_BCC-ESM1_historical_r1i1p1f1_gn_185001-201412.nc (64.95 MB) in 8.27 seconds (7.85 MB/s) from aims3.llnl.gov 2024-05-15 07:04:25,862 UTC [134535] INFO Downloaded /users/username/climate_data/cmip5/output1/BCC/bcc-csm1-1/historical/mon/atmos/Amon/r1i1p1/v1/tas_Amon_bcc-csm1-1_historical_r1i1p1_185001-201212.nc (64.15 MB) in 13.63 seconds (4.71 MB/s) from aims3.llnl.gov 2024-05-15 07:04:25,870 UTC [134535] INFO Downloaded 129.2 MB in 13.67 seconds (9.45 MB/s) 2024-05-15 07:04:25,870 UTC [134535] INFO Successfully downloaded all requested files. 2024-05-15 07:04:25,871 UTC [134535] INFO Using the Dask basic scheduler. 2024-05-15 07:04:25,871 UTC [134535] INFO Running 5 tasks using 5 processes 2024-05-15 07:04:25,956 UTC [144507] INFO Starting task map/tas in process [144507] 2024-05-15 07:04:25,956 UTC [144522] INFO Starting task timeseries/tas_amsterdam in process [144522] 2024-05-15 07:04:25,957 UTC [144534] INFO Starting task timeseries/tas_global in process [144534] 2024-05-15 07:04:26,049 UTC [134535] INFO Progress: 3 tasks running, 2 tasks waiting for ancestors, 0/5 done 2024-05-15 07:04:26,457 UTC [144534] WARNING Long name changed from 'Grid-Cell Area for Atmospheric Variables' to 'Grid-Cell Area for Atmospheric Grid Variables' (for file /users/username/climate_data/CMIP6/CMIP/BCC/BCC-ESM1/1pctCO2/r1i1p1f1/fx/areacella/gn/v20190613/areacella_fx_BCC-ESM1_1pctCO2_r1i1p1f1_gn.nc) 2024-05-15 07:04:26,461 UTC [144507] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:26,856 UTC [144522] INFO Extracting data for Amsterdam, Noord-Holland, Nederland (52.3730796 °N, 4.8924534 °E) 2024-05-15 07:04:27,081 UTC [144507] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:27,085 UTC [144534] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:40,666 UTC [144507] INFO Successfully completed task map/tas (priority 1) in 0:00:14.709864 2024-05-15 07:04:40,805 UTC [134535] INFO Progress: 2 tasks running, 2 tasks waiting for ancestors, 1/5 done 2024-05-15 07:04:40,813 UTC [144547] INFO Starting task map/script1 in process [144547] 2024-05-15 07:04:40,821 UTC [144547] INFO Running command ['/LUMI_TYKKY_D1Npoag/miniconda/envs/env1/bin/python', '/LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/diag_scripts/examples/diagnostic.py', '/users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/map/script1/settings.yml'] 2024-05-15 07:04:40,822 UTC [144547] INFO Writing output to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/work/map/script1 2024-05-15 07:04:40,822 UTC [144547] INFO Writing plots to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/plots/map/script1 2024-05-15 07:04:40,822 UTC [144547] INFO Writing log to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/map/script1/log.txt 2024-05-15 07:04:40,822 UTC [144547] INFO To re-run this diagnostic script, run: cd /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/map/script1; MPLBACKEND="Agg" /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/bin/python /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/diag_scripts/examples/diagnostic.py /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/map/script1/settings.yml 2024-05-15 07:04:40,906 UTC [134535] INFO Progress: 3 tasks running, 1 tasks waiting for ancestors, 1/5 done 2024-05-15 07:04:47,225 UTC [144522] INFO Extracting data for Amsterdam, Noord-Holland, Nederland (52.3730796 °N, 4.8924534 °E) 2024-05-15 07:04:47,308 UTC [144534] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:47,697 UTC [144534] INFO Successfully completed task timeseries/tas_global (priority 4) in 0:00:21.738941 2024-05-15 07:04:47,845 UTC [134535] INFO Progress: 2 tasks running, 1 tasks waiting for ancestors, 2/5 done 2024-05-15 07:04:48,053 UTC [144522] INFO Generated PreprocessorFile: /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/preproc/timeseries/tas_amsterdam/MultiModelMean_historical_Amon_tas_1850-2000.nc 2024-05-15 07:04:48,058 UTC [144522] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:48,228 UTC [144522] INFO Successfully completed task timeseries/tas_amsterdam (priority 3) in 0:00:22.271045 2024-05-15 07:04:48,346 UTC [134535] INFO Progress: 1 tasks running, 1 tasks waiting for ancestors, 3/5 done 2024-05-15 07:04:48,358 UTC [144558] INFO Starting task timeseries/script1 in process [144558] 2024-05-15 07:04:48,364 UTC [144558] INFO Running command ['/LUMI_TYKKY_D1Npoag/miniconda/envs/env1/bin/python', '/LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/diag_scripts/examples/diagnostic.py', '/users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/timeseries/script1/settings.yml'] 2024-05-15 07:04:48,365 UTC [144558] INFO Writing output to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/work/timeseries/script1 2024-05-15 07:04:48,365 UTC [144558] INFO Writing plots to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/plots/timeseries/script1 2024-05-15 07:04:48,365 UTC [144558] INFO Writing log to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/timeseries/script1/log.txt 2024-05-15 07:04:48,365 UTC [144558] INFO To re-run this diagnostic script, run: cd /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/timeseries/script1; MPLBACKEND="Agg" /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/bin/python /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/diag_scripts/examples/diagnostic.py /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/timeseries/script1/settings.yml 2024-05-15 07:04:48,447 UTC [134535] INFO Progress: 2 tasks running, 0 tasks waiting for ancestors, 3/5 done 2024-05-15 07:04:54,019 UTC [144547] INFO Maximum memory used (estimate): 0.4 GB 2024-05-15 07:04:54,021 UTC [144547] INFO Sampled every second. It may be inaccurate if short but high spikes in memory consumption occur. 2024-05-15 07:04:55,174 UTC [144547] INFO Successfully completed task map/script1 (priority 0) in 0:00:14.360271 2024-05-15 07:04:55,366 UTC [144558] INFO Maximum memory used (estimate): 0.4 GB 2024-05-15 07:04:55,368 UTC [144558] INFO Sampled every second. It may be inaccurate if short but high spikes in memory consumption occur. 2024-05-15 07:04:55,566 UTC [134535] INFO Progress: 1 tasks running, 0 tasks waiting for ancestors, 4/5 done 2024-05-15 07:04:56,958 UTC [144558] INFO Successfully completed task timeseries/script1 (priority 2) in 0:00:08.599797 2024-05-15 07:04:57,072 UTC [134535] INFO Progress: 0 tasks running, 0 tasks waiting for ancestors, 5/5 done 2024-05-15 07:04:57,072 UTC [134535] INFO Successfully completed all tasks. 2024-05-15 07:04:57,134 UTC [134535] INFO Wrote recipe with version numbers and wildcards to: file:///users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/recipe_python_filled.yml 2024-05-15 07:04:57,399 UTC [134535] INFO Wrote recipe output to: file:///users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/index.html 2024-05-15 07:04:57,399 UTC [134535] INFO Ending the Earth System Model Evaluation Tool at time: 2024-05-15 07:04:57 UTC 2024-05-15 07:04:57,400 UTC [134535] INFO Time for running the recipe was: 0:00:48.332409 2024-05-15 07:04:57,756 UTC [134535] INFO Maximum memory used (estimate): 2.5 GB 2024-05-15 07:04:57,757 UTC [134535] INFO Sampled every second. It may be inaccurate if short but high spikes in memory consumption occur. 2024-05-15 07:04:57,759 UTC [134535] INFO Removing `preproc` directory containing preprocessed data 2024-05-15 07:04:57,759 UTC [134535] INFO If this data is further needed, then set `remove_preproc_dir` to `false` in your user configuration file 2024-05-15 07:04:57,782 UTC [134535] INFO Run was successful
On Gadi with esmvaltool-workflow you will see the wrapper has run esmvaltool in a
PBS job for you, when complete you can find the output in
/scratch/nf33/$USER/esmvaltool_outputs/. In the run folder, the main_log would
be the terminal output of the command. This recipe won’t complete as it needs internet
connection to search for the location.
We will modify this recipe later so that it completes, for now you will likely see the below in your log file.
Error output
ERROR [2488385] Program terminated abnormally, see stack trace below for more information: multiprocessing.pool.RemoteTraceback: """ Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connection.py", line 196, in _new_conn sock = connection.create_connection( ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/util/connection.py", line 85, in create_connection raise err File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/util/connection.py", line 73, in create_connection sock.connect(sa) OSError: [Errno 101] Network is unreachable The above exception was the direct cause of the following exception: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 789, in urlopen response = self._make_request( ^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 490, in _make_request raise new_e File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 466, in _make_request self._validate_conn(conn) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 1095, in _validate_conn conn.connect() File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connection.py", line 615, in connect self.sock = sock = self._new_conn() ^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connection.py", line 211, in _new_conn raise NewConnectionError( urllib3.exceptions.NewConnectionError: <urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable The above exception was the direct cause of the following exception: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/adapters.py", line 667, in send resp = conn.urlopen( ^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 873, in urlopen return self.urlopen( ^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 873, in urlopen return self.urlopen( ^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 843, in urlopen retries = retries.increment( ^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/util/retry.py", line 519, in increment raise MaxRetryError(_pool, url, reason) from reason # type: ignore[arg-type] ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ urllib3.exceptions.MaxRetryError: HTTPSConnectionPool(host='nominatim.openstreetmap.org', port=443): Max retries exceeded with url: /search?q=Amsterdam&format=json&limit=1 (Caused by NewConnectionError('<urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable')) During handling of the above exception, another exception occurred: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/adapters.py", line 482, in _request resp = self.session.get(url, timeout=timeout, headers=headers) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/sessions.py", line 602, in get return self.request("GET", url, **kwargs) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/sessions.py", line 589, in request resp = self.send(prep, **send_kwargs) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/sessions.py", line 703, in send r = adapter.send(request, **kwargs) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/adapters.py", line 700, in send raise ConnectionError(e, request=request) requests.exceptions.ConnectionError: HTTPSConnectionPool(host='nominatim.openstreetmap.org', port=443): Max retries exceeded with url: /search?q=Amsterdam&format=json&limit=1 (Caused by NewConnectionError('<urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable')) During handling of the above exception, another exception occurred: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/multiprocessing/pool.py", line 125, in worker result = (True, func(*args, **kwds)) ^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 816, in _run_task output_files = task.run() ^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 264, in run self.output_files = self._run(input_files) ^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/__init__.py", line 684, in _run product.apply(step, self.debug) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/__init__.py", line 492, in apply self.cubes = preprocess(self.cubes, step, ^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/__init__.py", line 401, in preprocess result.append(_run_preproc_function(function, item, settings, ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/__init__.py", line 346, in _run_preproc_function return function(items, **kwargs) ^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/_regrid.py", line 403, in extract_location geolocation = geolocator.geocode(location) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/geocoders/nominatim.py", line 297, in geocode return self._call_geocoder(url, callback, timeout=timeout) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/geocoders/base.py", line 368, in _call_geocoder result = self.adapter.get_json(url, timeout=timeout, headers=req_headers) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/adapters.py", line 472, in get_json resp = self._request(url, timeout=timeout, headers=headers) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/adapters.py", line 494, in _request raise GeocoderUnavailable(message) geopy.exc.GeocoderUnavailable: HTTPSConnectionPool(host='nominatim.openstreetmap.org', port=443): Max retries exceeded with url: /search?q=Amsterdam&format=json&limit=1 (Caused by NewConnectionError('<urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable')) """ The above exception was the direct cause of the following exception: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_main.py", line 533, in run fire.Fire(ESMValTool()) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/fire/core.py", line 143, in Fire component_trace = _Fire(component, args, parsed_flag_args, context, name) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/fire/core.py", line 477, in _Fire component, remaining_args = _CallAndUpdateTrace( ^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/fire/core.py", line 693, in _CallAndUpdateTrace component = fn(*varargs, **kwargs) ^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_main.py", line 413, in run self._run(recipe, session) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_main.py", line 455, in _run process_recipe(recipe_file=recipe, session=session) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_main.py", line 130, in process_recipe recipe.run() File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_recipe/recipe.py", line 1095, in run self.tasks.run(max_parallel_tasks=self.session['max_parallel_tasks']) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 738, in run self._run_parallel(address, max_parallel_tasks) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 782, in _run_parallel _copy_results(task, running[task]) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 805, in _copy_results task.output_files, task.products = future.get() ^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/multiprocessing/pool.py", line 774, in get raise self._value geopy.exc.GeocoderUnavailable: HTTPSConnectionPool(host='nominatim.openstreetmap.org', port=443): Max retries exceeded with url: /search?q=Amsterdam&format=json&limit=1 (Caused by NewConnectionError('<urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable')) INFO [2488385] If you have a question or need help, please start a new discussion on https://github.com/ESMValGroup/ESMValTool/discussions If you suspect this is a bug, please open an issue on https://github.com/ESMValGroup/ESMValTool/issues To make it easier to find out what the problem is, please consider attaching the files run/recipe_*.yml and run/main_log_debug.txt from the output directory.
Pro tip: ESMValTool search paths
You might wonder how ESMValTool was able find the recipe file, even though it’s not in your working directory. All the recipe paths printed from
esmvaltool recipes listare relative to ESMValTool’s installation location. This is where ESMValTool will look if it cannot find the file by following the path from your working directory.
Investigating the log messages
Let’s dissect what’s happening here.
Output files and directories
After the banner and general information, the output starts with some important locations.
- Did ESMValTool use the right config file?
- What is the path to the example recipe?
- What is the main output folder generated by ESMValTool?
- Can you guess what the different output directories are for?
- ESMValTool creates two log files. What is the difference?
Answers
- The config file should be the one we edited in the previous episode, something like
/home/<username>/.esmvaltool/config-user.ymlor~/esmvaltool_tutorial/config-user.yml.- ESMValTool found the recipe in its installation directory, something like
/home/users/username/mambaforge/envs/esmvaltool/bin/esmvaltool/recipes/examples/or if you are using a pre-installed module on a server, something like/apps/jasmin/community/esmvaltool/ESMValTool_<version> /esmvaltool/recipes/examples/recipe_python.yml, where<version>is the latest release.- ESMValTool creates a time-stamped output directory for every run. In this case, it should be something like
recipe_python_YYYYMMDD_HHMMSS. This folder is made inside the output directory specified in the previous episode:~/esmvaltool_tutorial/esmvaltool_output.- There should be four output folders:
plots/: this is where output figures are stored.preproc/: this is where pre-processed data are stored.run/: this is where esmvaltool stores general information about the run, such as log messages and a copy of the recipe file.work/: this is where output files (not figures) are stored.- The log files are:
main_log.txtis a copy of the command-line outputmain_log_debug.txtcontains more detailed information that may be useful for debugging.
Debugging: No ‘preproc’ directory?
If you’re missing the preproc directory, then your
config-user.ymlfile has the valueremove_preproc_dirset totrue(this is used to save disk space). Please set this value tofalseand run the recipe again.
After the output locations, there are two main sections that can be distinguished in the log messages:
- Creating tasks
- Executing tasks
Analyse the tasks
List all the tasks that ESMValTool is executing for this recipe. Can you guess what this recipe does?
Answer
Just after all the ‘creating tasks’ and before ‘executing tasks’, we find the following line in the output:
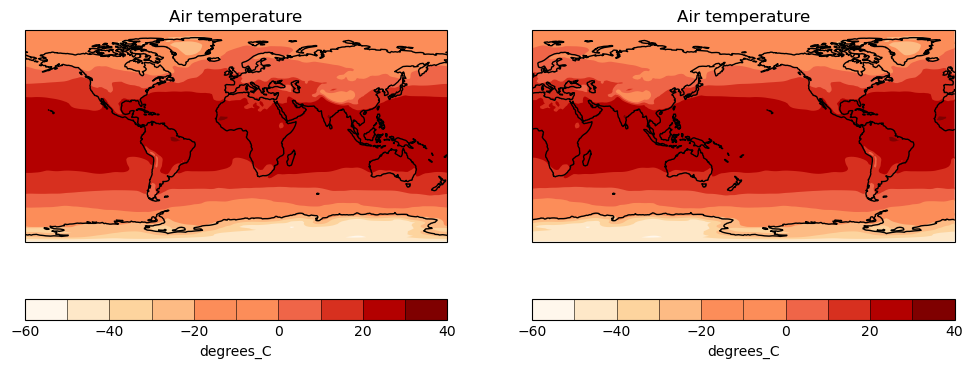
[134535] INFO These tasks will be executed: map/tas, timeseries/tas_global, timeseries/script1, map/script1, timeseries/tas_amsterdamSo there are three tasks related to timeseries: global temperature, Amsterdam temperature, and a script (tas: near-surface air temperature). And then there are two tasks related to a map: something with temperature, and again a script.
Examining the recipe file
To get more insight into what is happening, we will have a look at the recipe
file itself. Use the following command to copy the recipe to your working
directory (eg. in \scratch\nf33\$USERNAME\)
esmvaltool recipes get examples/recipe_python.yml
Now you should see the recipe file in your working directory (type ls to
verify). Use VS Code to open this file, you should be able to open from your
explorer panel:
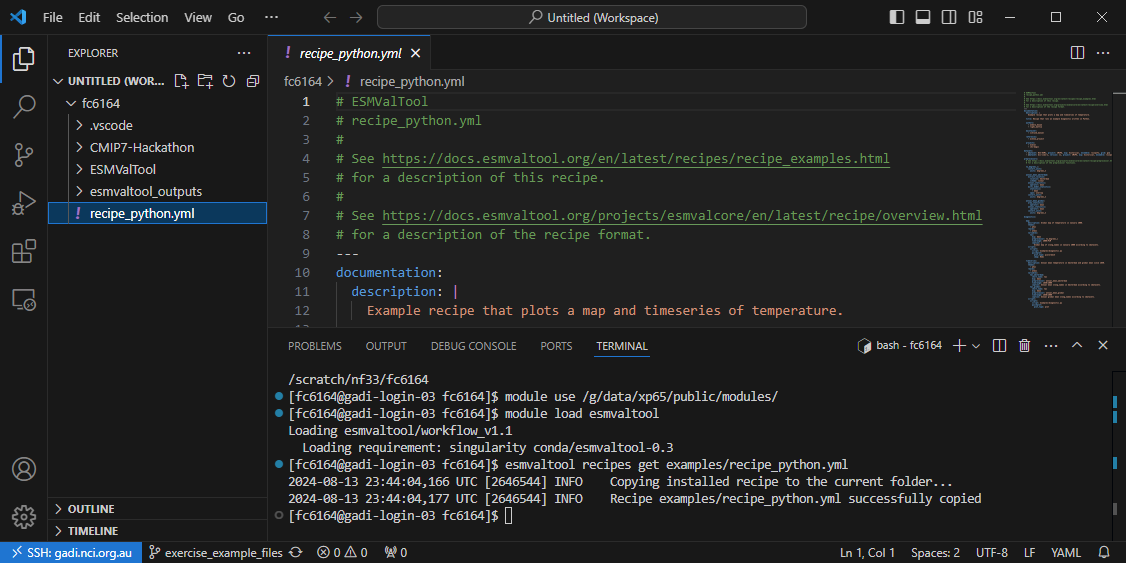
For reference, you can also view the recipe by unfolding the box below.
recipe_python.yml
# ESMValTool # recipe_python.yml # # See https://docs.esmvaltool.org/en/latest/recipes/recipe_examples.html # for a description of this recipe. # # See https://docs.esmvaltool.org/projects/esmvalcore/en/latest/recipe/overview.html # for a description of the recipe format. --- documentation: description: | Example recipe that plots a map and timeseries of temperature. title: Recipe that runs an example diagnostic written in Python. authors: - andela_bouwe - righi_mattia maintainer: - schlund_manuel references: - acknow_project projects: - esmval - c3s-magic datasets: - {dataset: BCC-ESM1, project: CMIP6, exp: historical, ensemble: r1i1p1f1, grid: gn} - {dataset: bcc-csm1-1, project: CMIP5, exp: historical, ensemble: r1i1p1} preprocessors: # See https://docs.esmvaltool.org/projects/esmvalcore/en/latest/recipe/preprocessor.html # for a description of the preprocessor functions. to_degrees_c: convert_units: units: degrees_C annual_mean_amsterdam: extract_location: location: Amsterdam scheme: linear annual_statistics: operator: mean multi_model_statistics: statistics: - mean span: overlap convert_units: units: degrees_C annual_mean_global: area_statistics: operator: mean annual_statistics: operator: mean convert_units: units: degrees_C diagnostics: map: description: Global map of temperature in January 2000. themes: - phys realms: - atmos variables: tas: mip: Amon preprocessor: to_degrees_c timerange: 2000/P1M caption: | Global map of {long_name} in January 2000 according to {dataset}. scripts: script1: script: examples/diagnostic.py quickplot: plot_type: pcolormesh cmap: Reds timeseries: description: Annual mean temperature in Amsterdam and global mean since 1850. themes: - phys realms: - atmos variables: tas_amsterdam: short_name: tas mip: Amon preprocessor: annual_mean_amsterdam timerange: 1850/2000 caption: Annual mean {long_name} in Amsterdam according to {dataset}. tas_global: short_name: tas mip: Amon preprocessor: annual_mean_global timerange: 1850/2000 caption: Annual global mean {long_name} according to {dataset}. scripts: script1: script: examples/diagnostic.py quickplot: plot_type: plot
Do you recognize the basic recipe structure that was introduced in episode 1?
- Documentation with relevant (citation) information
- Datasets that should be analysed
- Preprocessors groups of common preprocessing steps
- Diagnostics scripts performing more specific evaluation steps
Analyse the recipe
Try to answer the following questions:
- Who wrote this recipe?
- Who should be approached if there is a problem with this recipe?
- How many datasets are analyzed?
- What does the preprocessor called
annual_mean_globaldo?- Which script is applied for the diagnostic called
map?- Can you link specific lines in the recipe to the tasks that we saw before?
- How is the location of the city specified?
- How is the temporal range of the data specified?
Answers
- The example recipe is written by Bouwe Andela and Mattia Righi.
- Manuel Schlund is listed as the maintainer of this recipe.
- Two datasets are analysed:
- CMIP6 data from the model BCC-ESM1
- CMIP5 data from the model bcc-csm1-1
- The preprocessor
annual_mean_globalcomputes an area mean as well as annual means- The diagnostic called
mapexecutes a script referred to asscript1. This is a python script namedexamples/diagnostic.py- There are two diagnostics:
mapandtimeseries. Under the diagnosticmapwe find two tasks:
- a preprocessor task called
tas, applying the preprocessor calledto_degrees_cto the variabletas.- a diagnostic task called
script1, applying the scriptexamples/diagnostic.pyto the preprocessed data (map/tas).Under the diagnostic
timeserieswe find three tasks:
- a preprocessor task called
tas_amsterdam, applying the preprocessor calledannual_mean_amsterdamto the variabletas.- a preprocessor task called
tas_global, applying the preprocessor calledannual_mean_globalto the variabletas.- a diagnostic task called
script1, applying the scriptexamples/diagnostic.pyto the preprocessed data (timeseries/tas_globalandtimeseries/tas_amsterdam).- The
extract_locationpreprocessor is used to get data for a specific location here. ESMValTool interpolates to the location based on the chosen scheme. Can you tell the scheme used here? For more ways to extract areas, see the Area operations page.- The
timerangetag is used to extract data from a specific time period here. The start time is01/01/2000and the span of time to calculate means is1 Monthgiven byP1M. For more options on how to specify time ranges, see the timerange documentation.
Pro tip: short names and variable groups
The preprocessor tasks in ESMValTool are called ‘variable groups’. For the diagnostic
timeseries, we have two variable groups:tas_amsterdamandtas_global. Both of them operate on the variabletas(as indicated by theshort_name), but they apply different preprocessors. For the diagnosticmapthe variable group itself is namedtas, and you’ll notice that we do not explicitly provide theshort_name. This is a shorthand built into ESMValTool.
Output files
Have another look at the output directory created by the ESMValTool run.
Which files/folders are created by each task?
Answer
- map/tas: creates
/preproc/map/tas, which contains preprocessed data for each of the input datasets, a file calledmetadata.ymldescribing the contents of these datasets and provenance information in the form of.xmlfiles.- timeseries/tas_global: creates
/preproc/timeseries/tas_global, which contains preprocessed data for each of the input datasets, ametadata.ymlfile and provenance information in the form of.xmlfiles.- timeseries/tas_amsterdam: creates
/preproc/timeseries/tas_amsterdam, which contains preprocessed data for each of the input datasets, plus a combinedMultiModelMean, ametadata.ymlfile and provenance files.- map/script1: creates
/run/map/script1with general information and a log of the diagnostic script run. It also creates/plots/map/script1/and/work/map/script1, which contain output figures and output datasets, respectively. For each output file, there is also corresponding provenance information in the form of.xml,.bibtexand.txtfiles.- timeseries/script1: creates
/run/timeseries/script1with general information and a log of the diagnostic script run. It also creates/plots/timeseries/script1and/work/timeseries/script1, which contain output figures and output datasets, respectively. For each output file, there is also corresponding provenance information in the form of.xml,.bibtexand.txtfiles.
Pro tip: diagnostic logs
When you run ESMValTool, any log messages from the diagnostic script are not printed on the terminal. But they are written to the
log.txtfiles in the folder/run/<diag_name>/log.txt.ESMValTool does print a command that can be used to re-run a diagnostic script. When you use this the output will be printed to the command line.
Modifying the example recipe
Let’s make a small modification to the example recipe. Notice that now that you have copied and edited the recipe, you can use in your working directory:
esmvaltool-workflow run recipe_python.yml
to refer to your local file rather than the default version shipped with ESMValTool.
Change your location
Modify and run the recipe to analyse the temperature for your another location. Change the
extract_locationprerpocessor to one that doesn’t require internet connectionSolution
In principle, you only have to replace the
extract_locationwithextract_pointpreprocessor function and use latitude and longitude to define location. in the preprocessor calledannual_mean_amsterdam. However, it is good practice to also replace all instances ofamsterdamwith the correct name of your location. Otherwise the log messages and output will be confusing. You are free to modify the names of preprocessors or diagnostics.In the
difffile below you will see the changes we have made to the file. The top 2 lines are the filenames and the lines like@@ -39,9 +39,9 @@represent the line numbers in the original and modified file, respectively. For more info on this format, see here.--- recipe_python.yml +++ recipe_python_sydney.yml @@ -39,10 +39,9 @@ preprocessors: convert_units: units: degrees_C - annual_mean_amsterdam: - extract_location: - location: Amsterdam + annual_mean_sydney: + extract_point: + latitude: -34 + longitude: 151 scheme: linear annual_statistics: operator: mean @@ -84,18 +83,18 @@ diagnostics: themes: - phys realms: - atmos variables: - tas_amsterdam: + tas_sydney: short_name: tas mip: Amon - preprocessor: annual_mean_amsterdam + preprocessor: annual_mean_sydney timerange: 1850/2000 - caption: Annual mean {long_name} in Amsterdam according to {dataset}. + caption: Annual mean {long_name} in Sydney according to {dataset}. tas_global: short_name: tas mip: Amon
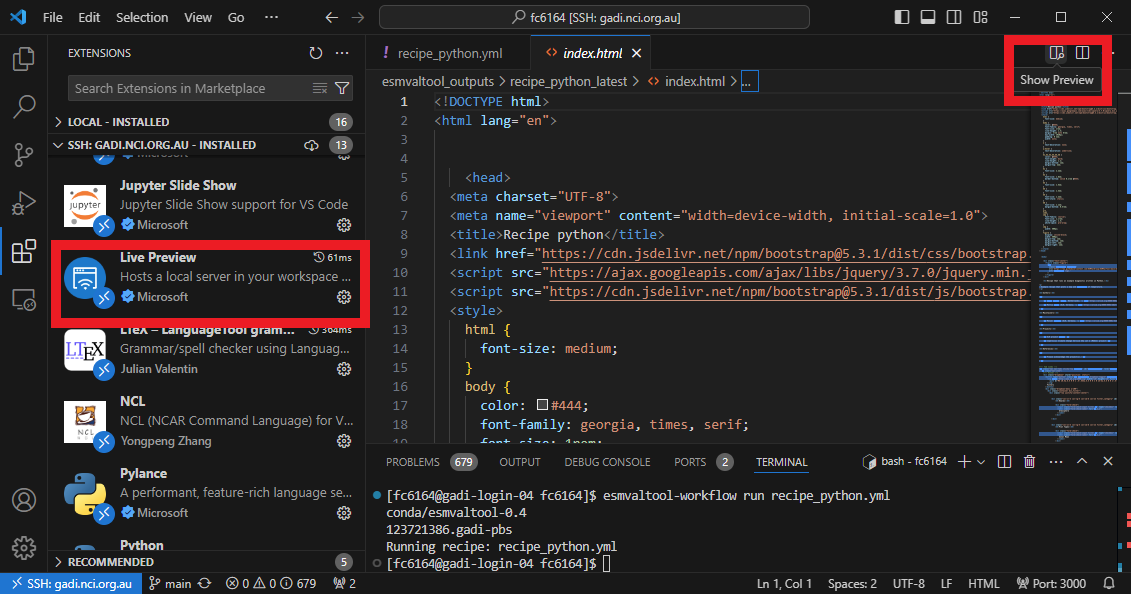
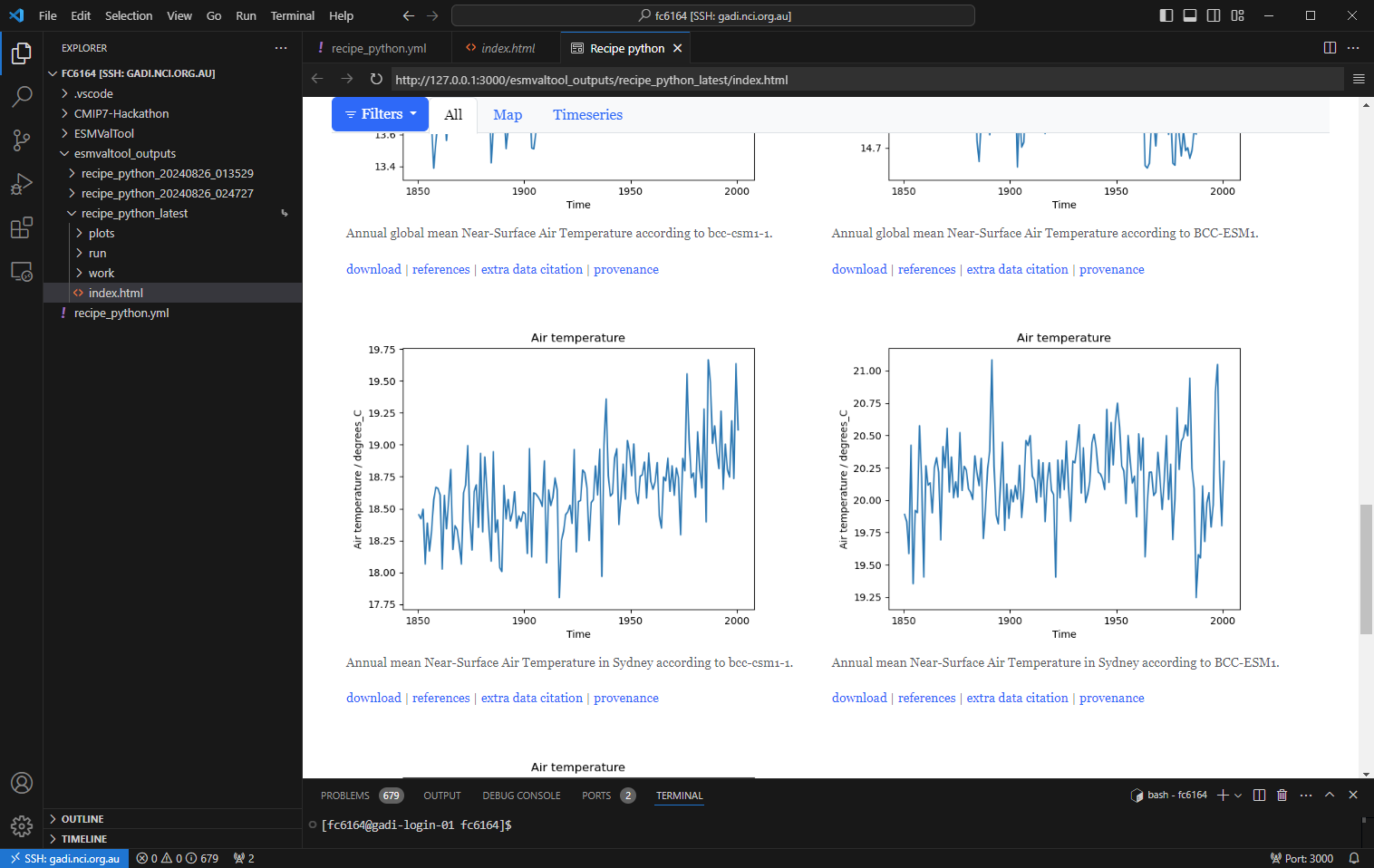
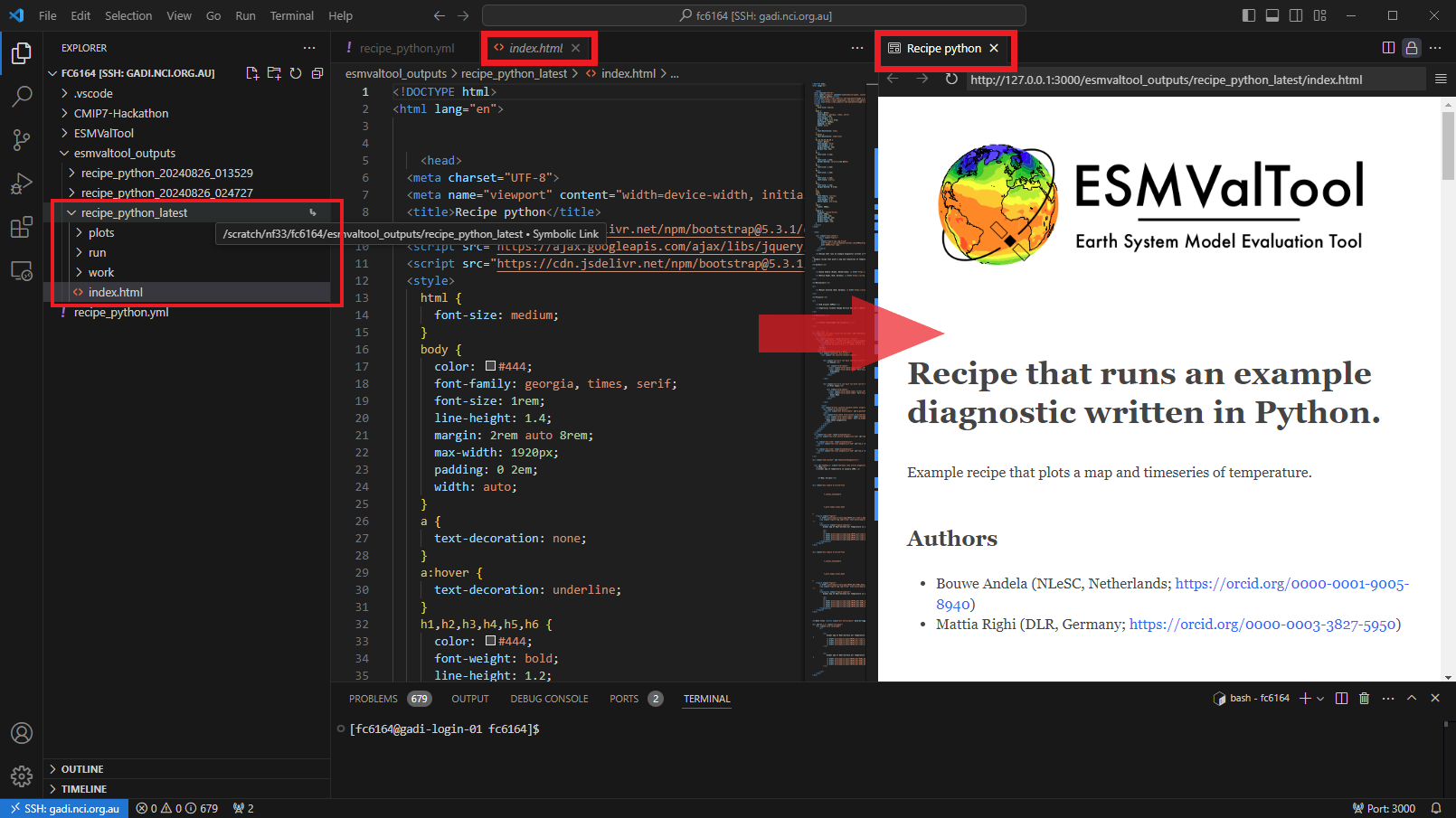
View the output
Now that the recipe runs we can look at the output. We recommend using VS Code with the “Live Preview” extension to view the html that is generated. When you open the html file, you will see the preview button appear in the top right.
Preview
You can see the output folder in explorer with the index.html file with a successful run. When you click on the preview button, the preview will appear to the right. You can also drag this across as a tab to use more of your screen to view.
HTML output
Key Points
ESMValTool recipes work ‘out of the box’ (if input data is available)
There are strong links between the recipe, log file, and output folders
Recipes can easily be modified to re-use existing code for your own use case
Supported data on NCI GADI
Overview
Teaching: 15 min
Exercises: 15 min
Compatibility:Questions
What data can I get on Gadi?
How can I access and find datasets?
Objectives
Gain knowledge of relevant Gadi projects for data
How observation data is organised for ESMValTool
Understanding download and CMORise functions available in ESMValTool
How observation data is organised for the ILAMB
Introduction
An advantage of using a supercomputer like Gadi at NCI, an ESGF node, is that a lot of data is already available which saves us from searching for and downloading large datasets that can’t be handled on other other computers.
What data can I get on Gadi?
Broadly, the datasets available which can be easily found and read in ESMValTool are:
- Observational data
- ERA5 data
- published CMIP6 data
- published CMIP5 data
What are the NCI projects I need to join?
On NCI, join relevant NCI projects to access that data. The NCI data catalogue can be searched for more information on the collections. Log into NCI with your NCI account to find and join the projects. These would have been checked when you ran the
check_hackathonset up.Data and NCI projects:
- You can check if you’re a member or join ct11 with this link.
The NCI data catalog entries with NCI project:
- ESMValTool observation data collection: ct11
- ESMValTool ERA5 Daily datasets: ct11
- ERA5 : rt52 and ERA5-Land: zz93
- CMIP6 replicas: oi10 and Australian published: fs38 and
- CMIP5 replicas: al33 and Australian: rr3
There is also the NCI project zv30 for CMIP7 collaborative development and evaluation which will be covered later in this episode.
Pro tip: Configuration file rootpaths
Remember the
config-user.ymlfile where we can set directories for ESMValTool to look for the data. This is an example from the Gadiesmvaltool-workflowuser configuration:config rootpaths
rootpath: CMIP6: [/g/data/oi10/replicas/CMIP6, /g/data/fs38/publications/CMIP6, /g/data/xp65/public/apps/esmvaltool/replicas/CMIP6] CMIP5: [/g/data/r87/DRSv3/CMIP5, /g/data/al33/replicas/CMIP5/combined, /g/data/rr3/publications/CMIP5/output1] CMIP3: /g/data/r87/DRSv3/CMIP3 CORDEX: [/g/data/rr3/publications/CORDEX/output, /g/data/al33/replicas/cordex/output] OBS: /g/data/ct11/access-nri/replicas/esmvaltool/obsdata-v2 OBS6: /g/data/ct11/access-nri/replicas/esmvaltool/obsdata-v2 obs4MIPs: [/g/data/ct11/access-nri/replicas/esmvaltool/obsdata-v2] ana4mips: [/g/data/ct11/access-nri/replicas/esmvaltool/obsdata-v2] native6: [/g/data/rt52/era5] ACCESS: /g/data/p73/archive/non-CMIP
ESMValTool Tiers
Observational datasets in ESMValTool are organised in tiers reflecting access restriction levels.
- Tier 1 Primarily Obs4MIPS where data is formatted, freely available and ready to use in ESMValTool.
- Tier 2 Data is freely available, CMORised datasets are available in ct11 in ESMValTool.
- Tier 3 These datasets have access rectrictions, licensing and acknowledgement may be required so direct access to the data cannot be provided. ACCESS-NRI can provide support to download and CMORise.
ERA5 in native6 and ERA5 daily in OBS6 Tier3
- native6-era5
The project native6
refers to a collection of datasets that can be read directly into CMIP6 format for
use in ESMValTool recipes. ESMValTool supports this with an extra facets file to map the variable names
across. This would have been added to your ~/.esmvaltool/extra_facets directory which is also used to fill
out default facet values and help find the data. See more information on
extra facets.
- ERA5 daily derived
The original hourly data from the “ERA5 hourly data on single levels” and “ERA5 hourly data on pressure levels”
collections have been transformed into daily means using the ESMValTool (v2.10) Python package.
These are Tier 3 datasets for OBS6. Variables available are:
'clt', 'fx', 'pr', 'prw', 'psl', 'rlds', 'rsds', 'rsdt',
'tas', 'tasmax', 'tasmin', 'tdps', 'ua', 'uas', 'vas'
What is the ESMValTool observation data collection?
We have created a collection of observation datasets that can be pulled directly into ESMValTool. The data has been CMORised, meaning they are netCDF files formatted to CF conventions and CMIP projects. There is a table of available Tier 1 and 2 data which can be found here or you can also expand the below:
Observation collection
long_name datasets name Ambient Aerosol Optical Thickness at 550nm ESACCI-AEROSOL, MODIS od550aer Surface Upwelling Shortwave Radiation CERES-EBAF, ESACCI-CLOUD, ISCCP-FH rsus Carbon Mass Flux out of Atmosphere Due to Net Biospheric Production on Land [kgC m-2 s-1] GCP2018, GCP2020 nbp Surface Temperature CFSR, ESACCI-LST, ESACCI-SST, HadISST, ISCCP-FH, NCEP-NCAR-R1 ts Daily Maximum Near-Surface Air Temperature E-OBS, NCEP-NCAR-R1 tasmax Omega (=dp/dt) NCEP-NCAR-R1 wap Surface Dissolved Inorganic Carbon Concentration OceanSODA-ETHZ dissicos Liquid Water Path ESACCI-CLOUD, MODIS lwp Surface Total Alkalinity OceanSODA-ETHZ talkos Eastward Wind CFSR, NCEP-NCAR-R1 ua Mole Fraction of N2O TCOM-N2O n2o Grid-Cell Area for Ocean Variables OceanSODA-ETHZ areacello Ambient Aerosol Optical Depth at 870nm ESACCI-AEROSOL od870aer Surface Carbonate Ion Concentration OceanSODA-ETHZ co3os Surface Upwelling Longwave Radiation CERES-EBAF, ESACCI-CLOUD, ISCCP-FH rlus Dissolved Oxygen Concentration CT2019, ESACCI-GHG, ESRL, GCP2018, GCP2020, Landschuetzer2016, Landschuetzer2020, OceanSODA-ETHZ, Scripps-CO2-KUM, WOA o2 Specific Humidity AIRS, AIRS-2-1, HALOE, JRA-25, NCEP-NCAR-R1, NOAA-CIRES-20CR hus TOA Outgoing Shortwave Radiation CERES-EBAF, ESACCI-CLOUD, ISCCP-FH, JRA-25, JRA-55, NCEP-NCAR-R1, NOAA-CIRES-20CR rsut Sea Water Salinity CALIPSO-GOCCP, ESACCI-LANDCOVER, ESACCI-SEA-SURFACE-SALINITY, PHC, WOA so Percentage Crop Cover ESACCI-LANDCOVER cropFrac Percentage of the Grid Cell Occupied by Land (Including Lakes) BerkeleyEarth sftlf Sea Surface Temperature ATSR, HadISST, WOA tos Total Dissolved Inorganic Silicon Concentration CFSR, GLODAP, HadISST, MOBO-DIC_MPIM, OSI-450-nh, OSI-450-sh, OceanSODA-ETHZ, PIOMAS, WOA si Daily Minimum Near-Surface Air Temperature E-OBS, NCEP-NCAR-R1 tasmin Dissolved Inorganic Carbon Concentration GLODAP, MOBO-DIC_MPIM, OceanSODA-ETHZ dissic Water Vapor Path ISCCP-FH, JRA-25, NCEP-DOE-R2, NCEP-NCAR-R1, NOAA-CIRES-20CR, SSMI, SSMI-MERIS prw Surface Downwelling Longwave Radiation CERES-EBAF, ISCCP-FH, JRA-55 rlds Geopotential Height CFSR, NCEP-NCAR-R1 zg Northward Wind CFSR, NCEP-NCAR-R1 va Relative Humidity AIRS-2-0, AIRS-2-1, NCEP-DOE-R2, NCEP-NCAR-R1 hur Tree Cover Percentage ESACCI-LANDCOVER treeFrac Percentage Cover by Shrub ESACCI-LANDCOVER shrubFrac Bare Soil Percentage Area Coverage ESACCI-LANDCOVER baresoilFrac Percentage Cloud Cover CALIOP, CALIPSO-GOCCP, CloudSat, ESACCI-CLOUD, ISCCP, JRA-25, JRA-55, MODIS, MODIS-1-0, NCEP-DOE-R2, NCEP-NCAR-R1, NOAA-CIRES-20CR, PATMOS-x cl Total Alkalinity GLODAP, OceanSODA-ETHZ talk Surface Upwelling Clear-Sky Shortwave Radiation CERES-EBAF, ESACCI-CLOUD rsuscs Mole Fraction of CH4 ESACCI-GHG, TCOM-CH4 ch4 Precipitation CRU, E-OBS, ESACCI-OZONE, GHCN, GPCC, GPCP-SG, ISCCP-FH, JRA-25, JRA-55, NCEP-DOE-R2, NCEP-NCAR-R1, NOAA-CIRES-20CR, PERSIANN-CDR, REGEN, SSMI, SSMI-MERIS, TRMM-L3, WFDE5, AGCD pr Ambient Fine Aerosol Optical Depth at 550nm ESACCI-AEROSOL od550lt1aer Sea Surface Salinity ESACCI-SEA-SURFACE-SALINITY, WOA sos Natural Grass Area Percentage ESACCI-LANDCOVER grassFrac Primary Organic Carbon Production by All Types of Phytoplankton Eppley-VGPM-MODIS intpp Eastward Near-Surface Wind CFSR uas Air Temperature AIRS, AIRS-2-1, BerkeleyEarth, CFSR, CRU, CowtanWay, E-OBS, GHCN-CAMS, GISTEMP, GLODAP, HadCRUT3, HadCRUT4, HadCRUT5, ISCCP-FH, Kadow2020, NCEP-DOE-R2, NCEP-NCAR-R1, NOAAGlobalTemp, OceanSODA-ETHZ, PHC, WFDE5, WOA ta Near-Surface Air Temperature BerkeleyEarth, CFSR, CRU, CowtanWay, E-OBS, GHCN-CAMS, GISTEMP, HadCRUT3, HadCRUT4, HadCRUT5, ISCCP-FH, Kadow2020, NCEP-NCAR-R1, NOAAGlobalTemp, WFDE5 tas Surface Downwelling Clear-Sky Longwave Radiation CERES-EBAF, JRA-55 rldscs Ambient Aerosol Absorption Optical Thickness at 550nm ESACCI-AEROSOL abs550aer Total Dissolved Inorganic Phosphorus Concentration WOA po4 Sea Level Pressure E-OBS, JRA-55, NCEP-NCAR-R1 psl Sea Water Potential Temperature PHC, WOA thetao CALIPSO Percentage Cloud Cover CALIPSO-GOCCP clcalipso Surface Aqueous Partial Pressure of CO2 Landschuetzer2016, Landschuetzer2020, OceanSODA-ETHZ spco2 Mass Concentration of Total Phytoplankton Expressed as Chlorophyll in Sea Water ESACCI-OC chl Surface pH OceanSODA-ETHZ phos TOA Outgoing Clear-Sky Longwave Radiation CERES-EBAF, ESACCI-CLOUD, ISCCP-FH, JRA-25, JRA-55, NCEP-NCAR-R1 rlutcs Total Column Ozone ESACCI-OZONE toz Near-Surface Relative Humidity NCEP-NCAR-R1 hurs Surface Downward Mass Flux of Carbon as CO2 [kgC m-2 s-1] GCP2018, GCP2020, Landschuetzer2016, OceanSODA-ETHZ fgco2 Atmosphere CO2 CT2019, ESRL, Scripps-CO2-KUM co2s pH GLODAP, OceanSODA-ETHZ ph Condensed Water Path MODIS, NOAA-CIRES-20CR clwvi Daily-Mean Near-Surface Wind Speed CFSR, NCEP-NCAR-R1 sfcWind Surface Downwelling Shortwave Radiation CERES-EBAF, ISCCP-FH rsds TOA Outgoing Clear-Sky Shortwave Radiation CERES-EBAF, ESACCI-CLOUD, ISCCP-FH, JRA-25, JRA-55, NCEP-NCAR-R1 rsutcs Total Cloud Cover Percentage CALIOP, CloudSat, ESACCI-CLOUD, ISCCP, JRA-25, JRA-55, MODIS, MODIS-1-0, NCEP-DOE-R2, NCEP-NCAR-R1, NOAA-CIRES-20CR, PATMOS-x clt Convective Cloud Area Percentage CALIOP, CALIPSO-GOCCP clc Northward Near-Surface Wind CFSR vas Surface Air Pressure CALIPSO-GOCCP, E-OBS, ISCCP-FH, JRA-55, NCEP-NCAR-R1 ps TOA Outgoing Longwave Radiation CERES-EBAF, ESACCI-CLOUD, ISCCP-FH, JRA-25, JRA-55, NCEP-NCAR-R1, NOAA-CIRES-20CR rlut Delta CO2 Partial Pressure Landschuetzer2016 dpco2 Surface Downwelling Clear-Sky Shortwave Radiation CERES-EBAF rsdscs TOA Incident Shortwave Radiation CERES-EBAF, ESACCI-CLOUD, ISCCP-FH rsdt Ice Water Path ESACCI-CLOUD clivi
ESMValTool data download and CMORise
ESMValTool has the capability to download and format certain observational datasets with data commands, see here for more detail and a table of datasets available to download and format. These are the download and format commands:
esmvaltool data download --config_file <path to config-user.yml> <dataset-name>
esmvaltool data format --config_file <path to config-user.yml> <dataset-name>
You will find the ESMValTool facet project for observational data can be OBS or OBS6
where OBS is CMIP5 format and OBS6 is CMIP6 format.
Finding data examples
Find data in recipe
Some facets can have glob patterns or wildcards for values. The facet
projectcannot be a wildcard, see reference.An example recipe that will use all CMIP6 datasets and all ensemble members which have a ‘historical’ experiment could look like this:
Solution
datasets: - project: CMIP6 exp: historical dataset: '*' institute: '*' ensemble: '*' grid: '*'
Find data using esmvalcore
This can be utilised through the esmvalcore API. To find all available datasets from ESGF which may not be available locally, set
search_esgfto always. This example looks for all ensembles for a dataset.Solution
from esmvalcore import Dataset from esmvalcore.config import CFG CFG['search_esgf'] = 'always' dataset_search = Dataset( short_name='tos', mip='Omon', project='CMIP6', exp='historical', dataset='ACCESS-ESM1-5', ensemble='*', grid='gn', ) ensemble_datasets = list(dataset_search.from_files()) ensemble_datasets
Find all available datasets for a variable in CMIP6
Find all datasets available for variable
tosin CMIP6 in concatenated experiments ‘historical’ and ‘ssp585’ for the time range 1850 to 2100.Solution
template = Dataset( short_name='tos', mip='Omon', activity='CMIP', institute='*', # facet req. to search locally project='CMIP6', exp= ['historical', 'ssp585'], dataset='*', # ensemble='*', grid='*', timerange='1850/2100' ) all_datasets = list(template.from_files()) all_datasets
What is ILAMB-Data?
The ILAMB community maintains a collection of reference datasets that have been carefully formatted following CF conventions. ACCESS-NRI hosts a replica of this ILAMB-data collection on NCI-Gadi as part of the ACCESS-NRI Replicated Datasets for Climate Model Evaluation NCI data collection, which can be accessed here. While we ensure this replica is regularly updated, the datasets were initially downloaded from primary sources and reformatted for use within the ILAMB framework. For specific reference information, please check the global attributes within the files.
See something wrong in a dataset? Have a suggestion? This collection is continually evolving and depends on community input. Please submit request for new observation datasets support on the ACCESS-Hive Forum. You can also track progress by following the ILAMB-Data GitHub repository or check out what the ILAMB community users are working on currently on the ILAMB Dataset Integration project board.
Observation collection
Albedo CERESed4.1, GEWEX.SRB Biomass ESACCI, GEOCARBON, NBCD2000, Saatchi2011, Thurner, USForest, XuSaatchi2021 Burned Area GFED4.1S Carbon Dioxide NOAA.Emulated, HIPPOAToM Diurnal Max Temperature CRU4.02 Diurnal Min Temperature CRU4.02 Diurnal Temperature Range CRU4.02 Ecosystem Respiration FLUXNET2015, FLUXCOM Evapotranspiration GLEAMv3.3a, MODIS, MOD16A2 Global Net Ecosystem Carbon Balance GCP, Hoffman Gross Primary Productivity FLUXNET2015, FLUXCOM, WECANN Ground Heat Flux CLASS Latent Heat FLUXNET2015, FLUXCOM, DOLCE, CLASS, WECANN Leaf Area Index AVHRR, AVH15C1, MODIS Methane FluxnetANN Net Ecosystem Exchange FLUXNET2015 Nitrogen Fixation Davies-Barnard Permafrost Brown2002, Obu2018 Precipitation CMAPv1904, FLUXNET2015, GPCCv2018, GPCPv2.3, CLASS Runoff Dai, LORA, CLASS Sensible Heat FLUXNET2015, FLUXCOM, CLASS, WECANN Snow Water Equivalent CanSISE Soil Carbon HWSD, NCSCDV22 Surface Air Temperature CRU4.02, FLUXNET2015 Surface Downward LW Radiation CERESed4.1, FLUXNET2015, GEWEX.SRB, WRMC.BSRN Surface Downward SW Radiation CERESed4.1, FLUXNET2015, GEWEX.SRB, WRMC.BSRN Surface Net LW Radiation CERESed4.1, FLUXNET2015, GEWEX.SRB, WRMC.BSRN Surface Net Radiation CERESed4.1, FLUXNET2015, GEWEX.SRB, WRMC.BSRN, CLASS Surface Net SW Radiation CERESed4.1, FLUXNET2015, GEWEX.SRB, WRMC.BSRN Surface Relative Humidity ERA5, CRU4.02 Surface Soil Moisture WangMao Surface Upward LW Radiation CERESed4.1, FLUXNET2015, GEWEX.SRB, WRMC.BSRN Surface Upward SW Radiation CERESed4.1, FLUXNET2015, GEWEX.SRB, WRMC.BSRN Terrestrial Water Storage Anomaly GRACE IOMB-DATA list
Alkalinity GLODAP2.2022 Anthropogenic DIC 1994-2007 Gruber, OCIM Chlorophyll GLODAP2.2022, SeaWIFS, MODISAqua Dissolved Inorganic Carbon GLODAP2.2022 Nitrate WOA2018, GLODAP2.2022 Oxygen WOA2018, GLODAP2.2022 Phosphate WOA2018, GLODAP2.2022 Salinity WOA2018, GLODAP2.2022 Silicate WOA2018, GLODAP2.2022 Temperature WOA2018, GLODAP2.2022 Vertical Temperature Gradient WOA2018, GLODAP2.2022
The CMIP7 collaborative development and evaluation project (zv30) on NCI-Gadi
The Australian CMIP7 community, supported by ACCESS-NRI, aims to establish a data space for effectively comparing and evaluating CMIP experiments in preparation for Australia’s forthcoming submission to CMIP7. This shared platform will serve as a collaborative hub, bringing together researchers and model developers to assess model outputs. It will enable comparisons with previous simulations and CMIP6 models, facilitating the real-time exchange of feedback. Additionally, this space will support iterative model improvement by providing a platform for testing and refining model configurations.
This collection is part of the zv30 project on NCI, managed by ACCESS-NRI. Similar to the NCI National data collections, users only have read access to this data. To share a dataset for model evaluation purposes, users must prepare the data according to CF conventions (i.e., CMORize the data) and submit a request to copy the dataset to the zv30 project. To do so, please contact Romain Beucher or Clare Richards at ACCESS-NRI.
If you have not done so already, please join the zv30 project
ZV30 collection in ESMValTool
ESMValTool-workflow on Gadi has been configured to be able to use this collection specifically and differentiate from the rest of the CMIP6 collections.
You can do this by specifying the project facet as
ZV30.In recipe
datasets: - project: ZV30 exp: piControl dataset: '*' institute: '*' ensemble: '*' grid: '*'
Key Points
There is supported data on Gadi to start with using ESMValTool and the ILAMB
Writing your own recipe
Overview
Teaching: 15 min
Exercises: 30 min
Compatibility:Questions
How do I create a new recipe?
Can I use different preprocessors for different variables?
Can I use different datasets for different variables?
How can I combine different preprocessor functions?
Can I run the same recipe for multiple ensemble members?
Objectives
Create a recipe with multiple preprocessors
Use different preprocessors for different variables
Run a recipe with variables from different datasets
Introduction
One of the key strengths of ESMValTool is in making complex analyses reusable and reproducible. But that doesn’t mean everything in ESMValTool needs to be complex. Sometimes, the biggest challenge is in keeping things simple. You probably know the ‘warming stripes’ visualization by Professor Ed Hawkins. On the site https://showyourstripes.info you can find the same visualization for many regions in the world.
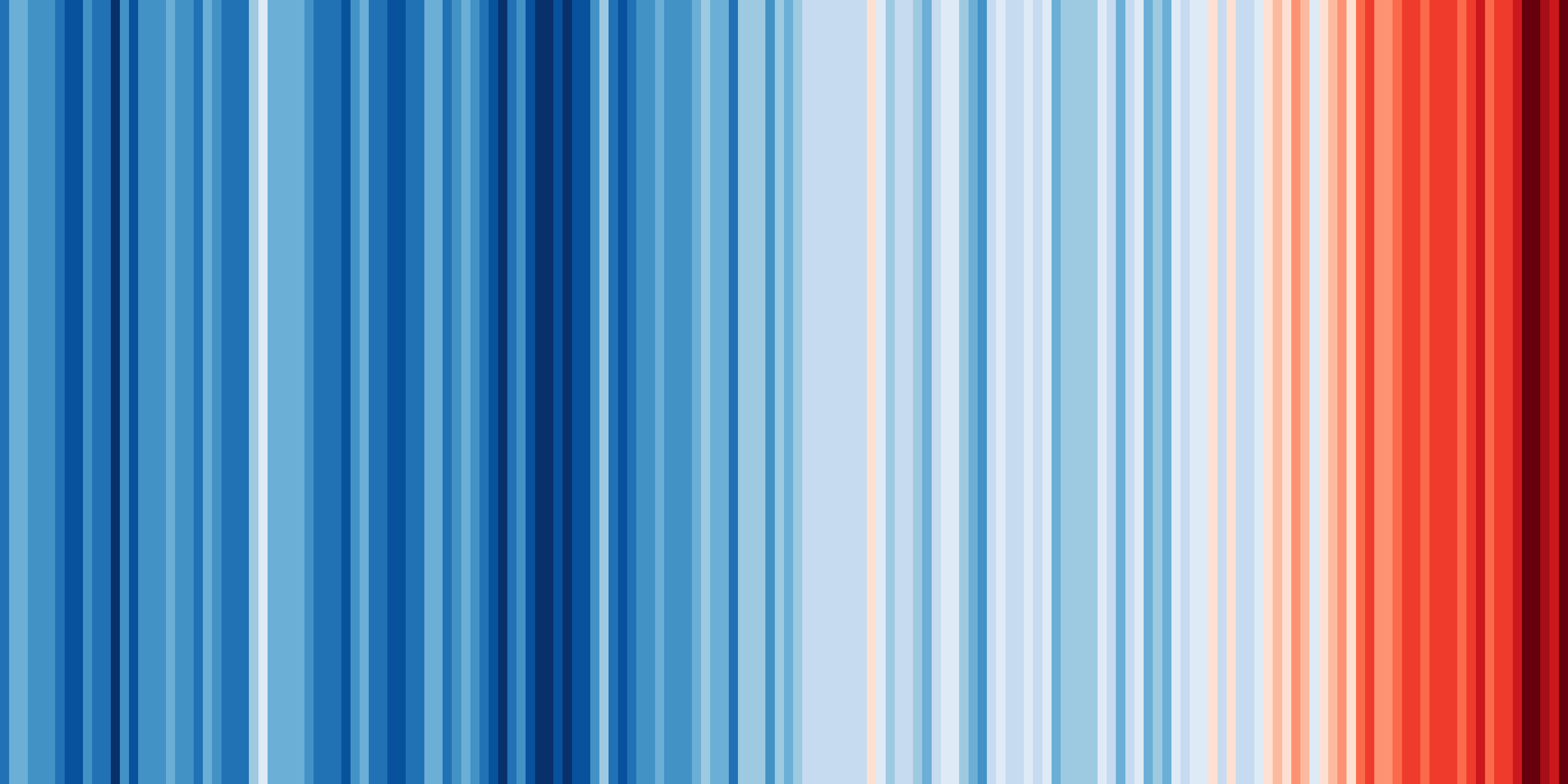 Shared by Ed Hawkins under a
Creative Commons 4.0 Attribution International licence. Source:
https://showyourstripes.info
Shared by Ed Hawkins under a
Creative Commons 4.0 Attribution International licence. Source:
https://showyourstripes.info
In this episode, we will reproduce and extend this functionality with ESMValTool. We have prepared a small Python script that takes a NetCDF file with timeseries data, and visualizes it in the form of our desired warming stripes figure.
As part of your setup when you ran check_hackathon you will have a clone of
this repo
in your scratch training space.
The diagnostic script that we will use is called warming_stripes.py and
can be found in your cloned Hackathon folder:
/scratch/nf33/$USER/CMIP7-Hackathon/exercises/WritingYourOwnRecipe.
You may also have a look at the contents, but it is not necessary to do so for this lesson.
We will write an ESMValTool recipe that takes some data, performs the necessary preprocessing, and then runs this Python script.
Drawing up a plan
Previously, we saw that running ESMValTool executes a number of tasks. What tasks do you think we will need to execute and what should each of these tasks do to generate the warming stripes?
Answer
In this episode, we will need to do the following two tasks:
- A preprocessing task that converts the gridded temperature data to a timeseries of global temperature anomalies
- A diagnostic task that calls our Python script, taking our preprocessed timeseries data as input.
Building a recipe from scratch
The easiest way to make a new recipe is to start from an existing one, and modify it until it does exactly what you need. However, in this episode we will start from scratch. This forces us to think about all the steps involved in processing the data. We will also deal with commonly occurring errors through the development of the recipe.
Remember the basic structure of a recipe, and notice that each component is extensively described in the documentation under the section, “Overview”:
This is the first place to look for help if you get stuck.
Create file and run on Gadi
Open VS Code with a remote SSH connection to Gadi with your /scratch/nf33/$USER folder in your workspace. Refer to VS Code setup Create a new file called
recipe_warming_stripes.ymlin your working directory for this exercise. Let’s add the standard header comments (these do not do anything), and a first description.# ESMValTool # recipe_warming_stripes.yml --- documentation: description: Reproducing Ed Hawkins' warming stripes visualization title: Reproducing Ed Hawkins' warming stripes visualization.Notice that
yamlalways requires two spaces indentation between the different levels. Save the file in VS Code withctrl + s.Reminder: how to run recipe
In the terminal, load the module to use ESMValTool on Gadi. If you don’t have a terminal open, the shortcut in VS Code is
Ctrl + `. Add the full path (eg. /scratch/nf33/$USER) to yourrecipe_warming_stripes.ymlin this when you run your recipe orcdto the directory. Also ensure that you are on the project nf33.switchproj nf33 module use /g/data/xp65/public/modules module load esmvaltool-workflow esmvaltool-workflow run --output_dir=/scratch/nf33/$USER/esmvaltool_outputs <dir_path>/recipe_warming_stripes.yml
If you try to run this, it would give an error. Below you see the last few lines of the error message.
...
yamale.yamale_error.YamaleError:
Error validating data '/home/users/username/esmvaltool_tutorial/recipe_warming_stripes.yml'
with schema
'/apps/jasmin/community/esmvaltool/miniconda3_py311_23.11.0-2/envs/esmvaltool/lib/python3.11/
site-packages/esmvalcore/_recipe/recipe_schema.yml'
documentation.authors: Required field missing
2024-05-27 13:21:23,805 UTC [41924] INFO
If you have a question or need help, please start a new discussion on
https://github.com/ESMValGroup/ESMValTool/discussions
If you suspect this is a bug, please open an issue on
https://github.com/ESMValGroup/ESMValTool/issues
To make it easier to find out what the problem is, please consider attaching the
files run/recipe_*.yml and run/main_log_debug.txt from the output directory.
We can use the log message above, to understand why ESMValTool failed. Here, this is because
we missed a required field with author names.
The text documentation.authors: Required field missing
tells us that. We see that ESMValTool always tries to validate the recipe
at an early stage. Note also the suggestion to open a GitHub issue if
you need help debugging the error message. This is something most
users do when they cannot understand the error or are not able to fix it
on their own.
Let’s add some additional information to the recipe. Open the recipe file again, and add an authors section below the description. ESMValTool expects the authors as a list, like so:
authors:
- lastname_firstname
To bypass a number of similar error messages, add a minimal diagnostics section below the documentation. The file should now look like:
# ESMValTool
# recipe_warming_stripes.yml
---
documentation:
description: Reproducing Ed Hawkins' warming stripes visualization
title: Reproducing Ed Hawkins' warming stripes visualization.
authors:
- doe_john
diagnostics:
dummy_diagnostic_1:
scripts: null
This is the minimal recipe layout that is required by ESMValTool. If we now run the recipe again, you will probably see the following error:
ValueError: Tag 'doe_john' does not exist in section
'authors' of /apps/jasmin/community/esmvaltool/ESMValTool_2.10.0/esmvaltool/config-references.yml
Pro tip: config-references.yml
The error message above points to a file named config-references.yml This is where ESMValTool stores all its citation information. To add yourself as an author, you will need to use and run ESMValTool in developer mode, then add your name in the form
lastname_firstnamein alphabetical order following the existing entries, under the# Development teamsection. The file used in this Gadi module doesn’t have editing permissions so use an existing author. See the List of authors section in the ESMValTool documentation for more information.
For now, let’s just use one of the existing references. Change the author field to
righi_mattia, who cannot receive enough credit for all the effort he put into
ESMValTool. If you now run the recipe, you would see the final message
ERROR No tasks to run!
Although there is no actual error in the recipe, ESMValTool assumes you mistakenly left out a variable name to process and alerts you with this error message.
Adding a dataset entry
Let’s add a datasets section.
Filling in the dataset keys
Use the paths specified in the configuration file to explore the data directory, and look at the explanation of the dataset entry in the ESMValTool documentation. For two datasets, write down the following properties:
- project
- variable (short name)
- CMIP table
- dataset (model name or obs/reanalysis dataset)
- experiment
- ensemble member
- grid
- start year
- end year
Answers
Here we have chosen a CMIP6 and CMIP5 ACCESS dataset.
key file 1 file 2 project CMIP6 CMIP5 short name tas tas CMIP table Amon Amon dataset ACCESS-ESM1-5 ACCESS1-0 experiment historical historical ensemble r1i1p1f1 r1i1p1 grid gn (native grid) N/A start year 1850 1850 end year 2014 2005 Note that the grid key is only required for CMIP6 data, and that the extent of the historical period has changed between CMIP5 and CMIP6.
Let us start with the ACCESS-ESM1-5 dataset and add a ‘datasets’ section to the recipe,
listing this single dataset, as shown below. Note that key fields such
as mip or start_year are included in the datasets section here but are part
of the diagnostic section in the recipe example seen in
Running your first recipe.
# ESMValTool
# recipe_warming_stripes.yml
---
documentation:
description: Reproducing Ed Hawkins' warming stripes visualization
title: Reproducing Ed Hawkins' warming stripes visualization.
authors:
- righi_mattia
datasets:
- {dataset: ACCESS-ESM1-5, project: CMIP6, mip: Amon, exp: historical,
ensemble: r1i1p1f1, grid: gn, start_year: 1850, end_year: 2014}
diagnostics:
dummy_diagnostic_1:
scripts: null
The recipe should run but produce the same message as in the previous case since we still have not included a variable to actually process. We have not included the short name of the variable in this dataset section because this allows us to reuse this dataset entry with different variable names later on. This is not really necessary for our simple use case, but it is common practice in ESMValTool.
Pro-tip: Automatically populating a recipe with all available datasets
You can select all available models for processing using
globpatterns or wildcards. Seen in Supported data on Gadi exercises on finding data.
Adding the preprocessor section
Above, we already described the preprocessing task that needs to convert the standard, gridded temperature data to a timeseries of temperature anomalies.
Defining the preprocessor
Have a look at the available preprocessors in the documentation. Write down
- Which preprocessor functions do you think we should use?
- What are the parameters that we can pass to these functions?
- What do you think should be the order of the preprocessors?
- A suitable name for the overall preprocessor
Solution
We need to calculate anomalies and global means. There is an
anomaliespreprocessor which takes in as arguments, a time period, a reference period, and whether or not to standardize the data. The global means can be calculated with thearea_statisticspreprocessor, which takes an operator as argument (in our case we want to compute themean).The default order in which these preprocessors are applied can be seen here:
area_statisticscomes beforeanomalies. If you want to change this, you can use thecustom_orderpreprocessor as described here. For this example, we will keep the default order.Let’s name our preprocessor
global_anomalies.
Add the following block to your recipe file between the datasets and diagnostics
block:
preprocessors:
global_anomalies:
area_statistics:
operator: mean
anomalies:
period: month
reference:
start_year: 1981
start_month: 1
start_day: 1
end_year: 2010
end_month: 12
end_day: 31
standardize: false
Completing the diagnostics section
We are now ready to finish our diagnostics section. Remember that we want to create two tasks: a preprocessor task, and a diagnostic task. To illustrate that we can also pass settings to the diagnostic script, we add the option to specify a custom colormap.
Fill in the blanks
Extend the diagnostics section in your recipe by filling in the blanks in the following template:
diagnostics: <... (suitable name for our diagnostic)>: description: <...> variables: <... (suitable name for the preprocessed variable)>: short_name: <...> preprocessor: <...> scripts: <... (suitable name for our python script)>: script: <full path to python script> colormap: <... choose from matplotlib colormaps>Solution
diagnostics: diagnostic_warming_stripes: description: visualize global temperature anomalies as warming stripes variables: global_temperature_anomalies: short_name: tas preprocessor: global_anomalies scripts: warming_stripes_script: script: /scratch/nf33/$USER/CMIP7-Hackathon/exercises/WritingYourOwnRecipe/warming_stripes.py colormap: 'bwr'
You should now be able to run the recipe from your working directory to get your own warming stripes.
esmvaltool-workflow run recipe_warming_stripes.yml
Find the plots in the plot directory of the output run eg.
/scratch/nf33/fc6164/esmvaltool_outputs/recipe_warming_latest/plots
└── diagnostic_warming_stripes
└── warming_stripes_script
└── CMIP6_ACCESS-ESM1-5_Amon_historical_r1i1p1f1_global_temperature_anomalies_gn_1850-2014.png
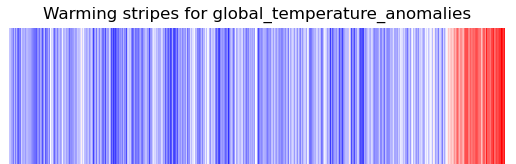
Note
For the purpose of simplicity in this episode, we have not added logging or provenance tracking in the diagnostic script. Once you start to develop your own diagnostic scripts and want to add them to the ESMValTool repositories, this will be required. Writing your own diagnostic script is discussed in a later episode.
Bonus exercises
Below are a few exercises to practice modifying an ESMValTool recipe. For your
reference, a copy of the recipe at this point can be found in the solution_recipes folder:
/scratch/nf33/$USER/CMIP7-Hackathon/exercises/Exercise2_files/solution_recipes.
Note the full path to the script will differ.
This will be the point of departure for each of the modifications we’ll make below.
An example of the modified recipes are also in this folder
Specific location selection
On showyourstripes.org, you can download stripes for specific locations. Here we show how this can be done with ESMValTool. Instead of the global mean, we can pick a location to plot the stripes for. Can you find a suitable preprocessor to do this?
Solution
You can use
extract_pointorextract_regionto select a location. We usedextract_regionfor Australia. A copy is called recipe_warming_stripes_local.yml and this is the difference from the previous recipe:--- recipe_warming_stripes.yml +++ recipe_warming_stripes_local.yml @@ -10,9 +10,11 @@ - {dataset: ACCESS-ESM1-5, project: CMIP6, mip: Amon, exp: historical, ensemble: r1i1p1f1, grid: gn, start_year: 1850, end_year: 2014} preprocessors: - global_anomalies: + aus_anomalies: + extract_region: + start_longitude: 110 + end_longitude: 160 + start_latitude: -45 + end_latitude: -9 area_statistics: operator: mean anomalies: period: month reference: @@ -29,9 +32,9 @@ diagnostics: diagnostic_warming_stripes: variables: - global_temperature_anomalies: + temperature_anomalies_aus: short_name: tas - preprocessor: global_anomalies + preprocessor: aus_anomalies scripts: warming_stripes_script: script: /scratch/nf33/$USER/CMIP7-Hackathon/exercises/WritingYourOwnRecipe/warming_stripes.py
Different time periods
Split the diagnostic in two with two different time periods for the same variable. You can choose the time periods yourself. In the example below, we have chosen the recent past and the 20th century and have used variable grouping.
Solution
This is the difference with the previous recipe:
--- recipe_warming_stripes_local.yml +++ recipe_warming_stripes_periods.yml @@ -7,7 +7,7 @@ datasets: - - {dataset: ACCESS-ESM1-5, project: CMIP6, mip: Amon, exp: historical, - ensemble: r1i1p1f1, grid: gn, start_year: 1850, end_year: 2014} + - {dataset: ACCESS-ESM1-5, project: CMIP6, mip: Amon, exp: historical, + ensemble: r1i1p1f1, grid: gn} preprocessors: anomalies_aus: @@ -31,9 +31,16 @@ diagnostics: diagnostic_warming_stripes: variables: - temperature_anomalies_aus: + temperature_anomalies_recent: short_name: tas preprocessor: anomalies_aus + start_year: 1950 + end_year: 2014 + temperature_anomalies_20th_century: + short_name: tas + preprocessor: anomalies_aus + start_year: 1900 + end_year: 1999 scripts: warming_stripes_script: script: /scratch/nf33/$USER/CMIP7-Hackathon/exercises/WritingYourOwnRecipe/warming_stripes.py
Different preprocessors
Now that you have different variable groups, we can also use different preprocessors. Add a second preprocessor to add another location of your choosing.
Solution
This is the difference with the previous recipe:
--- recipe_warming_stripes_periods.yml +++ recipe_warming_stripes_multiple_locations.yml @@ -19,7 +19,7 @@ end_latitude: -9 area_statistics: operator: mean - anomalies: + anomalies: &anomalies period: month reference: start_year: 1981 @@ -29,18 +29,24 @@ end_month: 12 end_day: 31 standardize: false + anomalies_sydney: + extract_point: + latitude: -34 + longitude: 151 + scheme: linear + anomalies: *anomalies diagnostics: diagnostic_warming_stripes: variables: - temperature_anomalies_recent: + temperature_anomalies_recent_aus: short_name: tas preprocessor: anomalies_amsterdam start_year: 1950 end_year: 2014 - temperature_anomalies_20th_century: + temperature_anomalies_20th_century_sydney: short_name: tas - preprocessor: anomalies_amsterdam + preprocessor: anomalies_sydney start_year: 1900 end_year: 1999 scripts:
Pro-tip: YAML anchors
If you want to avoid retyping the arguments used in your preprocessor, you can use YAML anchors as seen in the
anomaliespreprocessor specifications in the recipe above.
Additional datasets
So far we have defined the datasets in the datasets section of the recipe. However, it’s also possible to add specific datasets only for specific variables or variable groups. Take a look at the documentation to learn about the
additional_datasetskeyword here, and add a second dataset only for one of the variable groups.Solution
This is the difference with the previous recipe:
--- recipe_warming_stripes_multiple_locations.yml +++ recipe_warming_stripes_additional_datasets.yml @@ -49,6 +49,8 @@ preprocessor: anomalies_sydney start_year: 1900 end_year: 1999 + additional_datasets: + - {dataset: ACCESS1-3, project: CMIP5, mip: Amon, exp: historical, ensemble: r1i1p1} scripts: warming_stripes_script: script: /scratch/nf33/$USER/CMIP7-Hackathon/exercises/WritingYourOwnRecipe/warming_stripes.py
Multiple ensemble members
You can choose data from multiple ensemble members for a model in a single line.
Solution
The
datasetsection allows you to choose more than one ensemble member Changes made are shown in the diff output below:--- recipe_warming_stripes.yml +++ recipe_warming_stripes_multiple_ensemble_members.yml @@ -10,7 +10,7 @@ - ensemble: r1i1p1f1, grid: gn, start_year: 1850, end_year: 2014} + ensemble: "r(1:2)i1p1f1", grid: gn, start_year: 1850, end_year: 2014}
Pro-tip: Concatenating datasets
Check out the section on a different way to use multiple ensemble members or even multiple experiments at Concatenating data corresponding to multiple facets.
Key Points
A recipe can work with different preprocessors at the same time.
The setting
additional_datasetscan be used to add a different dataset.Variable groups are useful for defining different settings for different variables.
Multiple ensemble members and experiments can be analysed in a single recipe through concatenation.
Writing your own diagnostic script
Overview
Teaching: 20 min
Exercises: 30 min
Compatibility:Questions
How do I write a new diagnostic in ESMValTool?
How do I use the preprocessor output in a Python diagnostic?
Objectives
Write a new Python diagnostic script.
Explain how a diagnostic script reads the preprocessor output.
Introduction
The diagnostic script is an important component of ESMValTool and it is where the scientific analysis or performance metric is implemented. With ESMValTool, you can adapt an existing diagnostic or write a new script from scratch. Diagnostics can be written in a number of open source languages such as Python, R, Julia and NCL but we will focus on understanding and writing Python diagnostics in this lesson.
In this lesson, we will explain how to find an existing diagnostic and run it. Also, we will work with the recipe recipe_python.yml and the diagnostic script diagnostic.py called by this recipe that we have seen in the lesson Running your first recipe.
Let’s get started!
Understanding an existing Python diagnostic
A clone of the ESMValTool repository should be available in your user folder in the nf33
scratch folder (/scratch/nf33/$USER/ESMValTool). If not, please make sure to run the
check_hackathon command after loading the esmvaltool-workflow module, check for any errors.
The folder ESMValTool contains the source code of the tool. We can find the
recipe recipe_python.yml and the python script diagnostic.py in these
directories:
- ESMValTool/esmvaltool/recipes/examples/recipe_python.yml
- ESMValTool/esmvaltool/diag_scripts/examples/diagnostic.py
Let’s have look at the code in diagnostic.py.
For reference, we show the diagnostic code in the dropdown box below.
There are four main sections in the script:
- A description i.e. the
docstring(line 1). - Import statements (line 2-16).
- Functions that implement our analysis (line 21-102).
- A typical Python top-level script i.e.
if __name__ == '__main__'(line 105-108).
diagnostic.py
1: """Python example diagnostic.""" 2: import logging 3: from pathlib import Path 4: from pprint import pformat 5: 6: import iris 7: 8: from esmvaltool.diag_scripts.shared import ( 9: group_metadata, 10: run_diagnostic, 11: save_data, 12: save_figure, 13: select_metadata, 14: sorted_metadata, 15: ) 16: from esmvaltool.diag_scripts.shared.plot import quickplot 17: 18: logger = logging.getLogger(Path(__file__).stem) 19: 20: 21: def get_provenance_record(attributes, ancestor_files): 22: """Create a provenance record describing the diagnostic data and plot.""" 23: caption = caption = attributes['caption'].format(**attributes) 24: 25: record = { 26: 'caption': caption, 27: 'statistics': ['mean'], 28: 'domains': ['global'], 29: 'plot_types': ['zonal'], 30: 'authors': [ 31: 'andela_bouwe', 32: 'righi_mattia', 33: ], 34: 'references': [ 35: 'acknow_project', 36: ], 37: 'ancestors': ancestor_files, 38: } 39: return record 40: 41: 42: def compute_diagnostic(filename): 43: """Compute an example diagnostic.""" 44: logger.debug("Loading %s", filename) 45: cube = iris.load_cube(filename) 46: 47: logger.debug("Running example computation") 48: cube = iris.util.squeeze(cube) 49: return cube 50: 51: 52: def plot_diagnostic(cube, basename, provenance_record, cfg): 53: """Create diagnostic data and plot it.""" 54: 55: # Save the data used for the plot 56: save_data(basename, provenance_record, cfg, cube) 57: 58: if cfg.get('quickplot'): 59: # Create the plot 60: quickplot(cube, **cfg['quickplot']) 61: # And save the plot 62: save_figure(basename, provenance_record, cfg) 63: 64: 65: def main(cfg): 66: """Compute the time average for each input dataset.""" 67: # Get a description of the preprocessed data that we will use as input. 68: input_data = cfg['input_data'].values() 69: 70: # Demonstrate use of metadata access convenience functions. 71: selection = select_metadata(input_data, short_name='tas', project='CMIP5') 72: logger.info("Example of how to select only CMIP5 temperature data:\n%s", 73: pformat(selection)) 74: 75: selection = sorted_metadata(selection, sort='dataset') 76: logger.info("Example of how to sort this selection by dataset:\n%s", 77: pformat(selection)) 78: 79: grouped_input_data = group_metadata(input_data, 80: 'variable_group', 81: sort='dataset') 82: logger.info( 83: "Example of how to group and sort input data by variable groups from " 84: "the recipe:\n%s", pformat(grouped_input_data)) 85: 86: # Example of how to loop over variables/datasets in alphabetical order 87: groups = group_metadata(input_data, 'variable_group', sort='dataset') 88: for group_name in groups: 89: logger.info("Processing variable %s", group_name) 90: for attributes in groups[group_name]: 91: logger.info("Processing dataset %s", attributes['dataset']) 92: input_file = attributes['filename'] 93: cube = compute_diagnostic(input_file) 94: 95: output_basename = Path(input_file).stem 96: if group_name != attributes['short_name']: 97: output_basename = group_name + '_' + output_basename 98: if "caption" not in attributes: 99: attributes['caption'] = input_file 100: provenance_record = get_provenance_record( 101: attributes, ancestor_files=[input_file]) 102: plot_diagnostic(cube, output_basename, provenance_record, cfg) 103: 104: 105: if __name__ == '__main__': 106: 107: with run_diagnostic() as config: 108: main(config)
What is the starting point of a diagnostic?
- Can you spot a function called
mainin the code above?- What are its input arguments?
- How many times is this function mentioned?
Solution
- The
mainfunction is defined in line 65 asmain(cfg).- The input argument to this function is the variable
cfg, a Python dictionary that holds all the necessary information needed to run the diagnostic script such as the location of input data and various settings. We will next parse thiscfgvariable in themainfunction and extract information as needed to do our analyses (e.g. in line 68).- The
mainfunction is called near the very end on line 108. So, it is mentioned twice in our code - once where it is called by the top-level Python script and second where it is defined.
The function run_diagnostic
The function
run_diagnostic(line 107) is called a context manager provided with ESMValTool and is the main entry point for most Python diagnostics.
Create a copy of the files for you to edit
You would already have a copy of the
recipe_python.ymlfrom the lesson Running your first recipe. Use the file you edited after you ranesmvaltool recipes get examples/recipe_python.ymlUse the edited file from the completion of the lesson.
Copy the file
diagnostic.pyto your working folder to keep the ones in the repo as templates unaltered while you can more easily find the files you are editing. Edit your recipe to point to your copy ofdiagnostic.py. Also, note the location for when you run your recipe.Solution
Example of your working folder:
/scratch/nf33/$USER/Exercise_writeDiagnostic/recipe_python.yml /scratch/nf33/$USER/Exercise_writeDiagnostic/diagnostic.pyIn your
recipe_python.yml, edit the path to the diagnostic script.script1: script: /scratch/nf33/$USER/Exercise_writeDiagnostic/diagnostic.py quickplot:When running the recipe you can run to the full path of your recipe if you are not in that directory:
esmvaltool-workflow run /scratch/nf33/$USER/Exercise_writeDiagnostic/recipe_python.yml
Preprocessor-diagnostic interface
In the previous exercise, we have seen that the variable cfg is the input
argument of the main function. The first argument passed to the diagnostic
via the cfg dictionary is a path to a file called settings.yml.
The ESMValTool documentation page provides an overview of what is in this file, see
Diagnostic script interfaces.
What information do I need when writing a diagnostic script?
Load the module in Gadi if you haven’t already. We know how to change the configuration settings before running a recipe. First we set the option
remove_preproc_dirtofalsein the configuration file, then run the reciperecipe_python.yml: (Or look at the output folder from your previous working run.)module use /g/data/xp65/public/modules module load esmvaltool-workflow esmvaltool-workflow run <your_working_folder>/recipe_python.yml
- Can you find one example of the file
settings.ymlin therundirectory?- Open the file
settings.ymland look at theinput_fileslist. It contains paths to some filesmetadata.yml. What information do you think is saved in those files?Solution
- One example of
settings.ymlcan be found in the directory: /scratch/nf33/[username]/esmvaltool_outputs/recipe_python_latest/run/map/script1/settings.yml- The
metadata.ymlfiles hold information about the preprocessed data. There is one file for each variable having detailed information on your data including project (e.g., CMIP6, CMIP5), dataset names (e.g., BCC-ESM1, CanESM2), variable attributes (e.g., standard_name, units), preprocessor applied and time range of the data. You can use all of this information in your own diagnostic.
Diagnostic shared functions
Looking at the code in diagnostic.py, we see that input_data is
read from the cfg dictionary (line 68). Now we can group the input_data
according to some criteria such as the model or experiment. To do so,
ESMValTool provides many functions such as select_metadata (line 71),
sorted_metadata (line 75), and group_metadata (line 79). As you can see
in line 8, these functions are imported from esmvaltool.diag_scripts.shared
that means these are shared across several diagnostics scripts. A list of
available functions and their description can be found in
The ESMValTool Diagnostic API reference.
Extracting information needed for analysis
We have seen the functions used for selecting, sorting and grouping data in the script. What do these functions do?
Solution
There is a statement after use of
select_metadata,sorted_metadataandgroup_metadatathat starts withlogger.info(lines 72, 76 and 82). These lines print output to the log files. In the previous exercise, we ran the reciperecipe_python.yml. If you look at the log filerecipe_python_#_#/run/map/script1/log.txtinesmvaltool_outputdirectory, you can see the output from each of these functions, for example:2023-06-28 12:47:14,038 [2548510] INFO diagnostic,106 Example of how to group and sort input data by variable groups from the recipe: {'tas': [{'alias': 'CMIP5', 'caption': 'Global map of {long_name} in January 2000 according to ' '{dataset}.\n', 'dataset': 'bcc-csm1-1', 'diagnostic': 'map', 'end_year': 2000, 'ensemble': 'r1i1p1', 'exp': 'historical', 'filename': '~/recipe_python_20230628_124639/preproc/map/tas/ CMIP5_bcc-csm1-1_Amon_historical_r1i1p1_tas_2000-P1M.nc', 'frequency': 'mon', 'institute': ['BCC'], 'long_name': 'Near-Surface Air Temperature', 'mip': 'Amon', 'modeling_realm': ['atmos'], 'preprocessor': 'to_degrees_c', 'product': ['output1', 'output2'], 'project': 'CMIP5', 'recipe_dataset_index': 1, 'short_name': 'tas', 'standard_name': 'air_temperature', 'start_year': 2000, 'timerange': '2000/P1M', 'units': 'degrees_C', 'variable_group': 'tas', 'version': 'v1'}, {'activity': 'CMIP', 'alias': 'CMIP6', 'caption': 'Global map of {long_name} in January 2000 according to ' '{dataset}.\n', 'dataset': 'BCC-ESM1', 'diagnostic': 'map', 'end_year': 2000, 'ensemble': 'r1i1p1f1', 'exp': 'historical', 'filename': '~/recipe_python_20230628_124639/preproc/map/tas/ CMIP6_BCC-ESM1_Amon_historical_r1i1p1f1_tas_gn_2000-P1M.nc', 'frequency': 'mon', 'grid': 'gn', 'institute': ['BCC'], 'long_name': 'Near-Surface Air Temperature', 'mip': 'Amon', 'modeling_realm': ['atmos'], 'preprocessor': 'to_degrees_c', 'project': 'CMIP6', 'recipe_dataset_index': 0, 'short_name': 'tas', 'standard_name': 'air_temperature', 'start_year': 2000, 'timerange': '2000/P1M', 'units': 'degrees_C', 'variable_group': 'tas', 'version': 'v20181214'}]}This is how we can access preprocessed data within our diagnostic.
Diagnostic computation
After grouping and selecting data, we can read individual attributes (such as filename)
of each item. Here, we have grouped the input data by variables,
so we loop over the variables (line 88). Following this is a call to the
function compute_diagnostic (line 93). Let’s look at the
definition of this function in line 42, where the actual analysis of the data is done.
Note that output from the ESMValCore preprocessor is in the form of NetCDF files.
Here, compute_diagnostic uses
Iris to read data
from a netCDF file and performs an operation squeeze to remove any dimensions
of length one. We can adapt this function to add our own analysis. As an example,
here we calculate the bias using the average of the data using Iris cubes.
def compute_diagnostic(filename):
"""Compute an example diagnostic."""
logger.debug("Loading %s", filename)
cube = iris.load_cube(filename)
logger.debug("Running example computation")
cube = iris.util.squeeze(cube)
# Calculate a bias using the average of data
cube.data = cube.core_data() - cube.core_data.mean()
return cube
iris cubes
Iris reads data from NetCDF files into data structures called cubes. The data in these cubes can be modified, combined with other cubes’ data or plotted.
Reading data using xarray
Alternately, you can use xarray to read the data instead of Iris.
Solution
First, import
xarraypackage at the top of the script as:import xarray as xrThen, change the
compute_diagnosticas:def compute_diagnostic(filename): """Compute an example diagnostic.""" logger.debug("Loading %s", filename) dataset = xr.open_dataset(filename) #do your analyses on the data here return datasetCaution: If you read data using xarray keep in mind to change accordingly the other functions in the diagnostic which are dealing at the moment with Iris cubes.
Reading data using the netCDF4 package
Yet another option to read the NetCDF file data is to use the netCDF-4 Python interface to the netCDF C library.
Solution
First, import the
netCDF4package at the top of the script as:import netCDF4Then, change
compute_diagnosticas:def compute_diagnostic(filename): """Compute an example diagnostic.""" logger.debug("Loading %s", filename) nc_data = netCDF4.Dataset(filename,'r') #do your analyses on the data here return nc_dataCaution: If you read data using netCDF4 keep in mind to change accordingly the other functions in the diagnostic which are dealing at the moment with Iris cubes.
Diagnostic output
Plotting the output
Often, the end product of a diagnostic script is a plot or figure. The Iris cube
returned from the compute_diagnostic function (line 93) is passed to the
plot_diagnostic function (line 102). Let’s have a look at the definition of
this function in line 52. This is where we would plug in our plotting routine in the
diagnostic script.
More specifically, the quickplot function (line 60) can be replaced with the
function of our choice. As can be seen, this function uses
**cfg['quickplot'] as an input argument. If you look at the diagnostic
section in the recipe recipe_python.yml, you see quickplot is a key
there:
script1:
script: <path_to_script diagnostic.py>
quickplot:
plot_type: pcolormesh
cmap: Reds
This way, we can pass arguments such as the type of
plot pcolormesh and the colormap cmap:Reds from the recipe to the
quickplot function in the diagnostic.
Passing arguments from the recipe to the diagnostic
Change the type of the plot and its colormap and inspect the output figure.
Solution
In the recipe
recipe_python.yml, you could changeplot_typeandcmap. As an example, we chooseplot_type: pcolorandcmap: BuGn:script1: script: <path_to_script diagnostic.py> quickplot: plot_type: pcolor cmap: BuGnThe plot can be found at path_to_recipe_output/plots/map/script1/png.
ESMValTool gallery
ESMValTool makes it possible to produce a wide array of plots and figures as seen in the gallery.
Saving the output
In our example, the function save_data in line 56 is used to save the Iris
cube. The saved files can be found under the work directory in a .nc format.
There is also the function save_figure in line 62 to save the plots under the
plot directory in a .png format (or preferred format specified in your
configuration settings). Again, you may choose your own method of saving the output.
## in diagnostic.py ##
55: # Save the data used for the plot
56: save_data(basename, provenance_record, cfg, cube)
..
61: # And save the plot
62: save_figure(basename, provenance_record, cfg)
You will see that they are imported from esmvaltool.diag_scripts.shared and
take arguments such as cfg so that they can be saved in the appropriate output location.
Recording the provenance
When developing a diagnostic script, it is good practice to record
provenance. To do so, we use the function get_provenance_record (line 100).
Let us have a look at the definition of this function in line 21 where we
describe the diagnostic data and plot. Using the dictionary record, it is
possible to add custom provenance to our diagnostics output.
Provenance is stored in the W3C PROV XML
format and also in an SVG file under the work and plot directory. For
more information, see recording provenance.
You will see that the record gets parsed as an argument in the saving outputs functions above.
Congratulations!
You now know the basic diagnostic script structure and some available tools for putting together your own diagnostics. Have a look at existing recipes and diagnostics in the repository for more examples of functions you can use in your diagnostics!
Key Points
ESMValTool provides helper functions to interface a Python diagnostic script with preprocessor output.
Existing diagnostics can be used as templates and modified to write new diagnostics.
Helper functions can be imported from
esmvaltool.diag_scripts.sharedand used in your own diagnostic script.
Use a Jupyter Notebook to run a recipe
Overview
Teaching: 10 min
Exercises: 20 min
Compatibility:Questions
How to load the
esmvaltoolmodule in ARE?How to view and run a recipe in a Jupyter Notebook?
How to run a single diagnostic or preprocessor task?
Objectives
Learn about the esmvalcore experimental API
View the Recipe output in a Jupyter Notebook
This episode shows us how we can use ESMValTool in a Jupyter notebook. We are using material from a short tutorial from EGU22 and the documentation which is a good place for further reference.
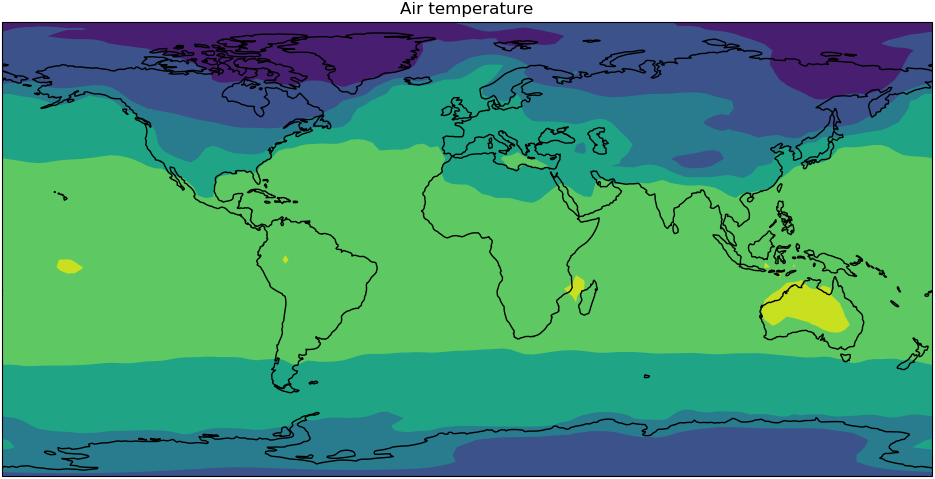
Start a session in ARE
Log in to ARE with your NCI account to start a JupyterLab session.
Refer to this ARE setup guide for more details.
Open the folder to your hackathon folder in nf33 where you can create a new notebook or use the
Intro_to_ESMValTool.ipynb notebook in CMIP7-Hackathon\exercises\Exercise4_files.
Let’s start by importing the tool and some other tools we can use later. Note that we are importing from esmvalcore and calling
it esmvaltool.
# Import the tool
import esmvalcore.experimental as esmvaltool
# Import tools for plotting
import matplotlib.pyplot as plt
import iris.quickplot
Finding a recipe
There is a utils
submodule we can use to find and get recipes. Call the get_all_recipes() function to get a
list of all available recipes from which you can use the find() method to return any matches. If you already know the
recipe you want you can use the get_recipe() function.
In Jupyter Notebook
all_recipes = esmvaltool.get_all_recipes() # all_recipes all_recipes.find('python')
Get a recipe
Let’s use the
examples/recipe_python.ymlfor this exercise, the documentation for it can be found here. Then see what’s in the recipe metadata.Solution
example_recipe = esmvaltool.get_recipe("examples/recipe_python.yml") example_recipeFor reading the recipe:
print(example_recipe.path.read_text())The
example_recipehere is a Recipe class with attributesdataandname, see the reference.example_recipe.name # 'Recipe python'
Pro tip: remember the command line?
This is similar to this function in the command line whihc copies the recipe to your directory.
>esmvaltool recipes get $recipeFile
Configuration in the notebook
We can look at the default user configuration file, ~/.esmvaltool/config-user.yml
by calling a CFG object as a dictionary. This gives us the ability to edit the settings.
The tool can automatically download the climate data files required to run a recipe for you.
You can check your download directory and output directory where your recipe runs will be saved.
This CFG object is from the config module in the ESMValCore API, for more details see here.
Call the
CFGobject and inspect the values.Solution
# call CFG object like this esmvaltool.CFGCheck output directory and change
Solution
Check this location is your
\scratch\nf33\$USERNAME\esmvaltool_outputs\print(CFG['output_dir']) # edit dir esmvaltool.CFG['output_dir'] = '/scratch/nf33/$USERNAME/esmvaltool_outputs'
Pro tip: Missing config file or load different config
Get configuration file
Rememeber that this command line copies and creates the default user configuration file in your home
.esmvaltoolfolder:esmvaltool config get-config-userLoad a different configuration file to use
# an example path to other configuration file esmvaltool.CFG.load_from_file('/home/189/fc6164/esmValTool/config-fc-copy.yml')
Running the recipe
Run the recipe and inspect the output.
Run
output = example_recipe.run() outputThis may take some time and you will see some logging messages as it runs
Inspect output
map/script1: ImageFile('CMIP5_bcc-csm1-1_Amon_historical_r1i1p1_tas_2000-P1M.png') ImageFile('CMIP6_BCC-ESM1_Amon_historical_r1i1p1f1_tas_gn_2000-P1M.png') DataFile('CMIP5_bcc-csm1-1_Amon_historical_r1i1p1_tas_2000-P1M.nc') DataFile('CMIP6_BCC-ESM1_Amon_historical_r1i1p1f1_tas_gn_2000-P1M.nc') timeseries/script1: ImageFile('tas_amsterdam_CMIP5_bcc-csm1-1_Amon_historical_r1i1p1_tas_1850-2000.png') ImageFile('tas_amsterdam_CMIP6_BCC-ESM1_Amon_historical_r1i1p1f1_tas_gn_1850-2000.png') ImageFile('tas_amsterdam_MultiModelMean_historical_Amon_tas_1850-2000.png') ImageFile('tas_global_CMIP5_bcc-csm1-1_Amon_historical_r1i1p1_tas_1850-2000.png') ImageFile('tas_global_CMIP6_BCC-ESM1_Amon_historical_r1i1p1f1_tas_gn_1850-2000.png') DataFile('tas_amsterdam_CMIP5_bcc-csm1-1_Amon_historical_r1i1p1_tas_1850-2000.nc') DataFile('tas_amsterdam_CMIP6_BCC-ESM1_Amon_historical_r1i1p1f1_tas_gn_1850-2000.nc') DataFile('tas_amsterdam_MultiModelMean_historical_Amon_tas_1850-2000.nc') DataFile('tas_global_CMIP5_bcc-csm1-1_Amon_historical_r1i1p1_tas_1850-2000.nc') DataFile('tas_global_CMIP6_BCC-ESM1_Amon_historical_r1i1p1f1_tas_gn_1850-2000.nc')
Pro tip: run a single Diagnostic
To run a single diagnostic, the name of the task can be passed as an argument to
run()output_1 = example_recipe.run('map/script1') output_1
Recipe output
The output can return the files as well as the image files and data files, also see the reference page.
Let’s look through this recipe output.
- Get the file paths.
- Look at one of the plots.
- Access and inspect the data used for the plots.
Solution
Print the file paths.
for result in output['map/script1']: print(result.path)Look at a plot from the list of plots.
plots = [f for f in output['timeseries/script1'] if isinstance(f, esmvaltool.recipe_output.ImageFile)] plots[-1]Load one of the preprocessed data files.
data_files = [f for f in output['map/script1'] if isinstance(f, esmvaltool.recipe_output.DataFile)] cube = data_files[0].load_iris()[0] cube
Use the loaded data to make your own plot in your notebook.
Solution
# Create plot iris.quickplot.contourf(cube) # Set the size of the figure plt.gcf().set_size_inches(12, 10) # Draw coastlines plt.gca().coastlines() # Show the resulting figure plt.show()
Key Points
ESMValTool can be run in a Jupyter Notebook
Access ImageFiles and DataFiles from the recipe run
Advanced Jupyter notebook
Overview
Teaching: 20 min
Exercises: 30 min
Compatibility:Questions
How to find data for ESMValTool in a Jupyter Notebook?
How to use preprocessor functions?
Objectives
Use the Dataset object
Import and use preprocessor functions
View and check the data
In this episode we will introduce the ESMValCore API in a jupyter notebook. This is reformatted from material from
this blog post
by Peter Kalverla. There’s also material from the example notebooks and the
API reference documentation.
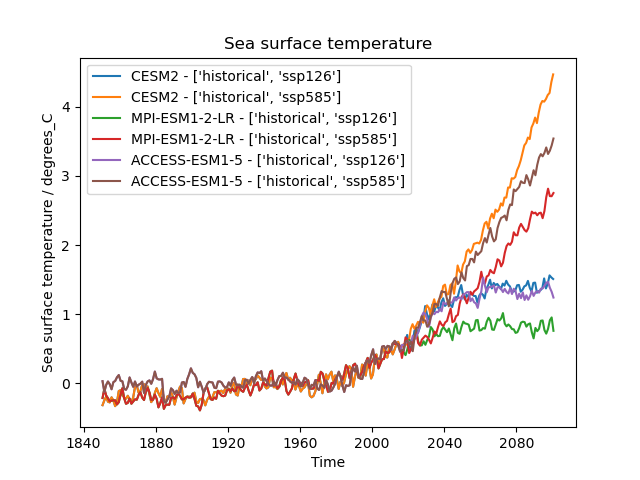
Start ARE session
Log in to ARE with your NCI account to start a JupyterLab session.
Refer to this ARE setup guide for more details.
Navigate to your hackathon folder /scratch/nf33/$USER/CMIP7-Hackathon/exercises/AdvancedJupyterNotebook
where you can find the example_easyipcc.ipynb notebook for this exercise.
Or you can create a new notebook in your workspace.
Find Datasets with facets
We have seen from running available recipes that ESMValTool is able to find data from facets that were given in
the recipe. We can use this in a Notebook, including filling out the facets for data definition.
To do this we will use the Dataset object from the API. Let’s look at this example.
from esmvalcore.dataset import Dataset
dataset = Dataset(
short_name='tos',
mip='Omon',
project='CMIP6',
exp='historical',
dataset='ACCESS-ESM1-5',
ensemble='r4i1p1f1',
grid='gn',
)
dataset.augment_facets()
print(dataset)
Pro tip: Augmented facets in the output
When running a recipe there is a
_filledrecipe in the output /run folder which augments the facets.Example recipe output folder
esmvaltool_output/flato13ipcc_figure914_CMIP6_20240729_043707/run ├── cmor_log.txt ├── fig09-14 ├── flato13ipcc_figure914_CMIP6_filled.yml * ├── flato13ipcc_figure914_CMIP6.yml ├── main_log_debug.txt ├── main_log.txt └── resource_usage.txt
Search available
Search from files locally with wildcard functionality
'*'to get the available datasets.
- How can you search for all available ensembles?
Solution
dataset_search = Dataset( short_name='tos', mip='Omon', project='CMIP6', exp='historical', dataset='ACCESS-ESM1-5', ensemble='*', grid='gn', ) ensemble_datasets = list(dataset_search.from_files()) print([ds['ensemble'] for ds in ensemble_datasets])There is also the ability to search on ESGF nodes and download. See reference for more details.
Add supplementary variables
Supplementary variables can be added to the
Datasetobject which can be used for certain preprocessors such as area statistics and weighting.
- Add the area file to this Dataset.
Solution
# Discard augmented facets as they will be different for areacello dataset = Dataset(**dataset.minimal_facets) # Add areacello as supplementary dataset dataset.add_supplementary(short_name='areacello', mip='Ofx') # Autocomplete and inspect dataset.augment_facets() print(dataset.summary())
Loading the data and inspect
# Before load, checks location of file print(dataset.files) cube = dataset.load() cubeOutput
sea_surface_temperature / (degC) (time: 1980; cell index along second dimension: 300; cell index along first dimension: 360) Dimension coordinates: time x - - cell index along second dimension - x - cell index along first dimension - - x Auxiliary coordinates: latitude - x x longitude - x x Cell measures: cell_area - x x Cell methods: 0 area: mean where sea 1 time: mean Attributes: Conventions 'CF-1.7 CMIP-6.2' activity_id 'CMIP' branch_method 'standard' branch_time_in_child 0.0 branch_time_in_parent -594980 cmor_version '3.4.0' data_specs_version '01.00.30' experiment 'all-forcing simulation of the recent past' experiment_id 'historical' external_variables 'areacello' forcing_index 1 frequency 'mon' further_info_url 'https://furtherinfo.es-doc.org/CMIP6.CSIRO.ACCESS-ESM1-5.historical.no ...' grid 'native atmosphere N96 grid (145x192 latxlon)' grid_label 'gn' initialization_index 1 institution 'Commonwealth Scientific and Industrial Research Organisation, Aspendale, ...' institution_id 'CSIRO' license 'CMIP6 model data produced by CSIRO is licensed under a Creative Commons ...' mip_era 'CMIP6' nominal_resolution '250 km' notes "Exp: ESM-historical; Local ID: HI-08; Variable: tos (['sst'])" parent_activity_id 'CMIP' parent_experiment_id 'piControl' parent_mip_era 'CMIP6' parent_source_id 'ACCESS-ESM1-5' parent_time_units 'days since 1850-1-1 00:00:00' parent_variant_label 'r1i1p1f1' physics_index 1 product 'model-output' realization_index 4 realm 'ocean' run_variant 'forcing: GHG, Oz, SA, Sl, Vl, BC, OC, (GHG = CO2, N2O, CH4, CFC11, CFC12, ...' source 'ACCESS-ESM1.5 (2019): \naerosol: CLASSIC (v1.0)\natmos: HadGAM2 (r1.1, ...' source_id 'ACCESS-ESM1-5' source_type 'AOGCM' sub_experiment 'none' sub_experiment_id 'none' table_id 'Omon' table_info 'Creation Date:(30 April 2019) MD5:40e9ef53d4d2ec9daef980b76f23d39a' title 'ACCESS-ESM1-5 output prepared for CMIP6' variable_id 'tos' variant_label 'r4i1p1f1' version 'v20200529'
Preprocessors
As mentioned in previous lessons, the idea of preprocessors are that they are a set of functions that can be applied in a centralised, documented and efficient way. There are a broad range of operations that are commonly done to input data before diagnostics or metrics are applied and can be done to all the datasets in a recipe consistently. See the documentation to read further.
Exercise: apply preprocessors using the API
See API reference to check the arguments for preprocessor functions. For this exercise, find;
- The global mean,
- Then anomalies which we can get monthly,
- Then aggregate annually for plotting and inspect the cube.
Solution
from esmvalcore.preprocessor import annual_statistics, anomalies, area_statistics # Set the reference period for anomalies reference_period = { "start_year": 1950, "start_month": 1, "start_day": 1, "end_year": 1979, "end_month": 12, "end_day": 31, } cube = area_statistics(cube, operator='mean') cube = anomalies(cube, reference=reference_period, period='month') cube = annual_statistics(cube, operator='mean') cube.convert_units('degrees_C') cubesea_surface_temperature / (degrees_C) (time: 165) Dimension coordinates: time x Auxiliary coordinates: year x Scalar coordinates: cell index along first dimension 179, bound=(0, 359) cell index along second dimension 149, bound=(0, 299) latitude 6.0 degrees_north, bound=(-78.0, 90.0) degrees_north longitude 179.9867706298828 degrees_east, bound=(0.0, 359.9735412597656) degrees_east Cell methods: 0 area: mean where sea 1 time: mean 2 latitude: longitude: mean 3 year: mean
Plot data
Iris has wrappers for matplotlib to plot the processed cubes. This is useful in a notebook to help develop your recipe with the esmvalcore preprocessors.
from iris import quickplot
quickplot.plot(cube)
Custom code
We have so far solely used ESMValCore, however, you can use your own custom code and
being in a Notebook means you can try straight away. Now, continue with other libraries
and make custom plots such as xarray.
import xarray as xr
da = xr.DataArray.from_iris(cube)
da.plot()
print(da)
Build workflow and diagnostic
Exercise - Easy IPCC plot for sea surface temperature
Let’s pull some of these bits together to build a diagnostic.
- Using the
Datasetobject, make a template which we can use to find multiple datasets we want to analyse together for variabletos.- The datasets being
"CESM2", "MPI-ESM1-2-LR", "ACCESS-ESM1-5"and experiments'ssp126', 'ssp585'with historical, iterate to build a list of datasets.- Apply the preprocessors to each dataset and plot the result
Solution
import cf_units import matplotlib.pyplot as plt from iris import quickplot from esmvalcore.config import CFG from esmvalcore.dataset import Dataset from esmvalcore.preprocessor import annual_statistics, anomalies, area_statistics # Settings for automatic ESGF search CFG['search_esgf'] = 'when_missing' # Declare common dataset facets template = Dataset( short_name='tos', mip='Omon', project='CMIP6', exp= '*', # We'll fill this below dataset='*', # We'll fill this below ensemble='r4i1p1f1', grid='gn', ) # Substitute data sources and experiments datasets = [] for dataset_id in ["CESM2", "MPI-ESM1-2-LR", "ACCESS-ESM1-5"]: for experiment_id in ['ssp126', 'ssp585']: dataset = template.copy(dataset=dataset_id, exp=['historical', experiment_id]) dataset.add_supplementary(short_name='areacello', mip='Ofx', exp='historical') dataset.augment_facets() datasets.append(dataset) # Set the reference period for anomalies reference_period = { "start_year": 1950, "start_month": 1, "start_day": 1, "end_year": 1979, "end_month": 12, "end_day": 31, } # (Down)load, pre-process, and plot the cubes for dataset in datasets: cube = dataset.load() cube = area_statistics(cube, operator='mean') cube = anomalies(cube, reference=reference_period, period='month') # notice 'month' cube = annual_statistics(cube, operator='mean') cube.convert_units('degrees_C') # Make sure all datasets use the same calendar for plotting tcoord = cube.coord('time') tcoord.units = cf_units.Unit(tcoord.units.origin, calendar='gregorian') # Plot quickplot.plot(cube, label=f"{dataset['dataset']} - {dataset['exp']}") # Show the plot plt.legend() plt.show()
Pro tip: Convert to recipe
We can use the helper to start making the recipe. A recipe can be used for reproducibility of an analysis. This list the datasets in a recipe format and we would then have to create the preprocessors and diagnostic script.
from esmvalcore.dataset import datasets_to_recipe import yaml for dataset in datasets: dataset.facets['diagnostic'] = 'easy_ipcc' print(yaml.safe_dump(datasets_to_recipe(datasets)))Output
datasets: - dataset: ACCESS-ESM1-5 exp: - historical - ssp126 - dataset: ACCESS-ESM1-5 exp: - historical - ssp585 - dataset: CESM2 exp: - historical - ssp126 - dataset: CESM2 exp: - historical - ssp585 - dataset: MPI-ESM1-2-LR exp: - historical - ssp126 - dataset: MPI-ESM1-2-LR exp: - historical - ssp585 diagnostics: easy_ipcc: variables: tos: ensemble: r4i1p1f1 grid: gn mip: Omon project: CMIP6 supplementary_variables: - exp: historical mip: Ofx short_name: areacello timerange: 1850/2100
Run through Minimal example notebook
Partly shown in the introduction episode. Find the example in your cloned hackathon folder:
CMIP7-Hackathon\exercises\IntroductionESMValTool\Minimal_example.ipynbThis notebook includes:
- Plot 2D field on a map
- Hovmoller Diagram
- Wind speed over Australia
- Air Potential Temperature (3D data) Transect
- Australian mean temperature timeseries
Exercise: Sea-ice area
Use observation data and 2 model datasets to show trends in sea-ice.
- Using variable
siconcwhich is a fraction percent(0-100)- Using datasets:
dataset:'ACCESS-ESM1-5', exp:'historical', ensemble:'r1i1p1f1', timerange:'1960/2010'dataset :'ACCESS-OM2', exp:'omip2', ensemble='r1i1p1f1', timerange:'0306/0366'- Using observations:
dataset:'NSIDC-G02202-sh', tier:'3', version:'4', timerange:'1979/2018'
- Extract Southern hemisphere
- Use only valid values (15 -100 %)
- Sum sea ice area which will be the fraction multiplied by cell area and summed
- Plot yearly minimum and maximum value
Solution notebook -
CMIP7-Hackathon/exercises/AdvancedJupyterNotebook/example_seaicearea.ipynb1. Define datasets:
from esmvalcore.dataset import Dataset obs = Dataset( short_name='siconc', mip='SImon', project='OBS6', type='reanaly', dataset='NSIDC-G02202-sh', tier='3', version='4', timerange='1979/2018', ) # Add areacello as supplementary dataset obs.add_supplementary(short_name='areacello', mip='Ofx') model = Dataset( short_name='siconc', mip='SImon', project='CMIP6', activity='CMIP', dataset='ACCESS-ESM1-5', ensemble='r1i1p1f1', grid='gn', exp='historical', timerange='1960/2010', institute = '*', ) om_facets={'dataset' :'ACCESS-OM2', 'exp':'omip2', 'activity':'OMIP', 'timerange':'0306/0366' } model.add_supplementary(short_name='areacello', mip='Ofx') model_om = model.copy(**om_facets)Tip: Check dataset files can be found
The observational dataset used is a Tier 3, so with some licensing restrictions. It is not directly accesible here. Check files can be found for all the datasets:
for ds in [model, model_om, obs]: print(ds['dataset'],' : ' ,ds.files) print(ds.supplementaries[0].files)This observation dataset does have a downloader and formatter with ESMValTool, you can use these data functions mentioned in the supported data lesson:
esmvaltool data download --config_file <path to config-user.yml> NSIDC-G02202-sh esmvaltool data format --config_file <path to config-user.yml> NSIDC-G02202-shFor this plot we can drop it for now. But you can also try to find and add another dataset. eg:
obs_other = Dataset( short_name='siconc', mip='*', project='OBS', type='*', dataset='*', tier='*', timerange='1979/2018' ) obs_other.files2. Use esmvalcore API preprocessors on the datasets and plot results
import iris import matplotlib.pyplot as plt from iris import quickplot from esmvalcore.preprocessor import ( mask_outside_range, extract_region, area_statistics, annual_statistics ) # om - at index 1 to offset years # drop observations that cannot be found load_data = [model, model_om] #, obs] # function to use for both min and max ['max','min'] def trends_seaicearea(min_max): plt.clf() for i,data in enumerate(load_data): cube = data.load() cube = mask_outside_range(cube, 15, 100) cube = extract_region(cube,0,360,-90,0) cube = area_statistics(cube, 'sum') cube = annual_statistics(cube, min_max) iris.util.promote_aux_coord_to_dim_coord(cube, 'year') cube.convert_units('km2') if i == 1: ## om years 306/366 apply offset cube.coord('year').points = [y + 1652 for y in cube.coord('year').points] label_name = data['dataset'] print(label_name, cube.shape) quickplot.plot(cube, label=label_name) plt.title(f'Trends in Sea-Ice {min_max.title()}ima') plt.ylabel('Sea-Ice Area (km2)') plt.legend() trends_seaicearea('min')
Key Points
API can be used as a helper to develop recipes
Preprocessors can be used in a Jupyter Notebook to check the output
Use
datasets_to_recipehelper to start making recipes
Running the ILAMB on Gadi
Overview
Teaching: 30 min
Exercises: 60 min
Compatibility:Questions
How do I run the ILAMB on NCI GADI?
Objectives
Understand how to load, configure and run the ILAMB using the ACCESS-NRI ILAMB-Workflow
What is the ILAMB?
The International Land Model Benchmarking (ILAMB) project is a model-data intercomparison and integration project designed to improve the performance of land models and, in parallel, improve the design of new measurement campaigns to reduce uncertainties associated with key land surface processes.
The purpose of the Quickstart Guide is to provide users of GADI with a streamlined process to rapidly run the International Land Model Benchmarking (ILAMB) system. ACCESS-NRI offers an already configured ILAMB module via the ILAMB-Workflow, enabling users to quickly initiate benchmarking tasks without the need for deployment. This guide is designed to help users efficiently begin evaluating land model outputs against observational datasets with minimal setup time.
How to cite the ILAMB?
Collier, N., Hoffman, F. M., Lawrence, D. M., Keppel-Aleks, G., Koven, C. D., Riley, W. J., et al. (2018). The International Land Model Benchmarking (ILAMB) system: Design, theory, and implementation. Journal of Advances in Modeling Earth Systems, 10, 2731–2754. https://doi.org/10.1029/2018MS001354
The ILAMB on NCI-Gadi
For NCI users, ACCESS-NRI is providing a conda environment with the latest version of ILAMB through project xp65..
module use /g/data/xp65/public/modules
module load ilamb-workflow
or
module use /g/data/xp65/public/modules
module load conda/access-med
To run the ILAMB, you need to execute the command ilamb-run with a number of arguments/files:
ilamb-run --config config.cfg --model_setup model_setup.txt --regions global
config.cfgdefines which observables and observational datasets will be comparedmodel_setup.txtdefines the paths of the models that will be compared
Below we explain how to setup the necessary directory structures and the example files mentioned above. For detailed information on the arguments of
ilamb-run, please consult the official ILAMB documentation.
Organising Data and Model Outputs for ILAMB Benchmarking
ILAMB requires files to be organized within a specific directory structure, consisting of DATA and MODELS directories. The DATA directory contains observational datasets, while the MODELS directory holds the output from the models you wish to benchmark. Adhering to this structure is essential for ILAMB to correctly locate and compare the datasets during the benchmarking process.
The following directory tree represents a typical ILAMB_ROOT setup for CMIP comparison on NCI/Gadi:
$ILAMB_ROOT/
|-- DATA -> /g/data/ct11/access-nri/replicas/ILAMB
|-- MODELS
|-- ACCESS-ESM1-5
| `-- piControl
| `-- r3i1p1f1
├── evspsbl.nc
├── hfds.nc
├── hfls.nc
├── hfss.nc
├── hurs.nc
├── pr.nc
├── rlds.nc
├── rlus.nc
├── rsds.nc
├── rsus.nc
├── tasmax.nc
├── tasmin.nc
├── tas.nc
└── tsl.nc
The top level of this directory structure is defined by the ILAMB_ROOT path, which should be set as an environment variable:
export ILAMB_ROOT=/path/to/your/ILAMB_ROOT/directory
By exporting this path as $ILAMB_ROOT, you ensure that the ILAMB can correctly locate the necessary directories and files during the benchmarking process.”
- the
DATAdirectory: this is where we keep the observational datasets each in a subdirectory bearing the name of the variable. - the
MODELdirectory: this directory can be populated with symbolic links to the model outputs.
Automating ILAMB Directory Structure Setup with ilamb-tree-generator
To simplify the setup of an ILAMB-ROOT directory tree, ACCESS-NRI offers a tool called ilamb-tree-generator, available within the ILAMB-Workflow through the access-med environment of the xp65 project.
The ilamb-tree-generator automates the creation of the necessary ILAMB directory structure. It efficiently generates symlinks to the ACCESS-NRI Replicated Datasets for Climate Model Evaluation and to the relevant sections of the model outputs. This automation helps ensure that your ILAMB benchmarking setup is correctly configured with minimal manual intervention.
To add model outputs, you can list them in a YAML file, formatted as follows:
datasets:
- {mip: CMIP, institute: CSIRO-ARCCSS, dataset: ACCESS-CM2, project: CMIP6, exp: piControl, ensemble: r3i1p1f1}
- {mip: CMIP, institute: CSIRO-ARCCSS, dataset: ACCESS-CM2, project: CMIP6, exp: piControl, ensemble: r3i1p2f1}
- {mip: CMIP, institute: CSIRO-ARCCSS, dataset: ACCESS-CM2, project: CMIP6, exp: piControl, ensemble: r3i1p3f1}
Once your YAML file is ready, you can run the tool from the command line to generate the directory structure:
ilamb-tree-generator --datasets models.yml --ilamb_root $ILAMB_ROOT
This command will automatically create the appropriate folders under the specified ILAMB_ROOT path, ensuring that your data is organized correctly for ILAMB benchmarking.”
Exercise
Copy the above to a models.yml file and try to run the
ilamb-tree-generator
ILAMB model selection: model_setup.txt
In the model_setup.txt, you can select all the model outputs that you want to compare.
Assuming you want to compare the three models that we used in ILAMB_ROOT/MODELS, you would need to create a model_setup.txt file wehere you define both the model labels and their paths:
# Model Name (used as label), ABSOLUTE/PATH/TO/MODELS or relative to $ILAMB_ROOT/ , Time Shift
piControl_r3i1p1f1, /scratch/nf33/yz9299/ILAMB-sep-2024/ILAMB_ROOT/MODELS/ACCESS-CM2/piControl/r3i1p1f1/, 1000, 1920
piControl_r3i1p2f1, /scratch/nf33/yz9299/ILAMB-sep-2024/ILAMB_ROOT/MODELS/ACCESS-CM2/piControl/r3i1p2f1/, 1000, 1920
piControl_r3i1p3f1, /scratch/nf33/yz9299/ILAMB-sep-2024/ILAMB_ROOT/MODELS/ACCESS-CM2/piControl/r3i1p3f1/, 1000, 1920
Since ILAMB require model-output data and observational data should have time overlap. In this case, our piControl data time-range is (1000-1080)
, and most of the observational data time range is (1900-2000), so we specify time shift in model_setup.txt from 1000 to 1920, make it comparable with observational data.
Configuring and Running a Benchmark Study with the ILAMB
ILAMB uses a config.cfg file as its configuration file to initiate a benchmark study. This file allows you to set up comparison sections and specify which variables from which datasets will be compared.
An example configuration file for ILAMB on Gadi might be named config.cfg. It could be used to compare your models with two variables from the radiation and energy cycle, as measured by the Clouds and the Earth’s Radiant Energy System (CERES) project:
This configuration file is used to define the comparison sections, variables, and observational datasets required for running ILAMB on Gadi. The file is organised with the following structure:
[h1:] Sections
[h2:] Variables
[] Observational Datasets
- Sections: Define the major comparison categories or groups within the benchmark study.
- Variables: Specify the particular variables that will be compared between model outputs and observational data.
- Observational Datasets: List the datasets used for comparison, detailing where ILAMB will source the observational data.
For further guidance on how to create and use configuration files, refer to the ILAMB Tutorial on Configure Files. You can also consult the ILAMB and IOMB dataset collections at ILAMB Datasets.
A minimal Example
[h1: Hydrology Cycle]
bgcolor = "#E6F9FF"
[h2: Evapotranspiration]
variable = "et"
alternate_vars = "evspsbl"
cmap = "Blues"
weight = 5
mass_weighting = True
[MODIS]
source = "DATA/evspsbl/MODIS/et_0.5x0.5.nc"
weight = 15
table_unit = "mm d-1"
plot_unit = "mm d-1"
relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02"
this example configuration file is set up for running ILAMB on Gadi and specifies details for comparing data related to the hydrology cycle. Here’s a breakdown of what each section does:
[h1: Hydrology Cycle]
bgcolor = "#E6F9FF"
- [h1: Hydrology Cycle]: This section defines a major comparison category called “Hydrology Cycle” and sets a background color for visualizations.
[h2: Evapotranspiration]
variable = "et"
alternate_vars = "evspsbl"
cmap = "Blues"
weight = 5
mass_weighting = True
- [h2: Evapotranspiration]: This subsection focuses on “Evapotranspiration” within the hydrology cycle.
variable: Specifies the main variable to compare, which is “et” (evapotranspiration).alternate_vars: Provides an alternate variable name “evspsbl” that might be used in the data.cmap: Sets the color map for plotting the data, here using shades of blue.weight: Assigns a weight of 5 to this variable in the comparisons.mass_weighting: Indicates that mass weighting should be applied (True).
[MODIS]
source = "DATA/evspsbl/MODIS/et_0.5x0.5.nc"
weight = 15
table_unit = "mm d-1"
plot_unit = "mm d-1"
relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02"
- [MODIS]: This section specifies details for the observational dataset related to MODIS.
source: Points to the file location of the MODIS dataset.weight: Assigns a weight of 15 to this dataset in the comparisons.table_unit: Defines the unit of measurement for the dataset, “mm d-1” (millimeters per day).plot_unit: Specifies the unit of measurement for plotting, also “mm d-1”.relationships: Lists other related datasets, such as precipitation and surface air temperature, indicating how they relate to the MODIS dataset.
Exercise: Adding a Second Observational Dataset to the ILAMB Configuration File
In this exercise, you will add a second observational dataset to your ILAMB configuration file. Follow these steps to integrate a new dataset,
[MOD16A2], into your existing configuration:
Open Your ILAMB Configuration File: Locate and open the ILAMB configuration file you are currently using.
- Identify the Section for Observational Datasets:
- Scroll to the section of the file where observational datasets are listed.
- Add the New Dataset:
- Insert the following block of code to include the
[MOD16A2]observational dataset:[MOD16A2] source = "DATA/evspsbl/MOD16A2/et.nc" weight = 15 table_unit = "mm d-1" plot_unit = "mm d-1" relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02"
- This entry specifies the details for the new dataset:
source: Path to the dataset file.weight: Weight assigned to this dataset for comparisons.table_unit: Unit of measurement used in tables.plot_unit: Unit of measurement used in plots.relationships: Lists other related datasets for comparison.- Save Your Changes: Make sure to save the configuration file after adding the new dataset.
Solution
# This configure file specifies comparison sections, variables and observational data for running ILAMB on Gadi. # See https://www.ilamb.org/doc/first_steps.html#configure-files for the ILAMB Tutorial that addesses Configure Files # See https://www.ilamb.org/datasets.html for the ILAMB and IOMB collections # Structure: # [h1:] Sections # [h2:] Variables # [] Observational Datasets #======================================================================================= [h1: Hydrology Cycle] bgcolor = "#E6F9FF" [h2: Evapotranspiration] variable = "et" alternate_vars = "evspsbl" cmap = "Blues" weight = 5 mass_weighting = True [MODIS] source = "DATA/evspsbl/MODIS/et_0.5x0.5.nc" weight = 15 table_unit = "mm d-1" plot_unit = "mm d-1" relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02" [MOD16A2] source = "DATA/evspsbl/MOD16A2/et.nc" weight = 15 table_unit = "mm d-1" plot_unit = "mm d-1" relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02" #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Exercise: Adding New Comparison Details to the ILAMB Configuration File
In this exercise, you will add a new section for “Latent Heat” to the ILAMB configuration file. Follow the steps below:
Open your existing ILAMB configuration file: Locate and open the configuration file you have been working with.
- Add a new subsection for Latent Heat:
- Scroll to the appropriate location in the file where you want to add the new details.
- Insert the following content to define the “Latent Heat” comparison:
[h2: Latent Heat] variable = "hfls" alternate_vars = "le" cmap = "Oranges" weight = 5 mass_weighting = True
- This section sets up a comparison for “Latent Heat,” specifying the variable, alternate names, color map, weight, and mass weighting.
- Add details for the FLUXCOM dataset:
- Below the “Latent Heat” subsection, add the following content to define the FLUXCOM dataset:
[FLUXCOM] source = "DATA/hfls/FLUXCOM/le.nc" land = True weight = 9 skip_iav = True
- This section specifies the source file for the FLUXCOM dataset, assigns a weight, indicates whether land data is included, and whether to skip inter-annual variability.
- Save your changes: Ensure that the file is saved with the new sections included.
Solution
# This configure file specifies comparison sections, variables and observational data for running ILAMB on Gadi. # See https://www.ilamb.org/doc/first_steps.html#configure-files for the ILAMB Tutorial that addesses Configure Files # See https://www.ilamb.org/datasets.html for the ILAMB and IOMB collections # Structure: # [h1:] Sections # [h2:] Variables # [] Observational Datasets #======================================================================================= [h1: Hydrology Cycle] bgcolor = "#E6F9FF" [h2: Evapotranspiration] variable = "et" alternate_vars = "evspsbl" cmap = "Blues" weight = 5 mass_weighting = True [MODIS] source = "DATA/evspsbl/MODIS/et_0.5x0.5.nc" weight = 15 table_unit = "mm d-1" plot_unit = "mm d-1" relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02" [MOD16A2] source = "DATA/evspsbl/MOD16A2/et.nc" weight = 15 table_unit = "mm d-1" plot_unit = "mm d-1" relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02" #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Latent Heat] variable = "hfls" alternate_vars = "le" cmap = "Oranges" weight = 5 mass_weighting = True [FLUXCOM] source = "DATA/hfls/FLUXCOM/le.nc" land = True weight = 9 skip_iav = True #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
A Comprehensive example
# This configure file specifies comparison sections, variables and observational data for running ILAMB on Gadi. # See https://www.ilamb.org/doc/first_steps.html#configure-files for the ILAMB Tutorial that addesses Configure Files # See https://www.ilamb.org/datasets.html for the ILAMB and IOMB collections # Structure: # [h1:] Sections # [h2:] Variables # [] Observational Datasets #======================================================================================= [h1: Hydrology Cycle] bgcolor = "#E6F9FF" [h2: Evapotranspiration] variable = "et" alternate_vars = "evspsbl" cmap = "Blues" weight = 5 mass_weighting = True [MODIS] source = "DATA/evspsbl/MODIS/et_0.5x0.5.nc" weight = 15 table_unit = "mm d-1" plot_unit = "mm d-1" relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02" [MOD16A2] source = "DATA/evspsbl/MOD16A2/et.nc" weight = 15 table_unit = "mm d-1" plot_unit = "mm d-1" relationships = "Precipitation/GPCPv2.3","SurfaceAirTemperature/CRU4.02" #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Latent Heat] variable = "hfls" alternate_vars = "le" cmap = "Oranges" weight = 5 mass_weighting = True [FLUXCOM] source = "DATA/hfls/FLUXCOM/le.nc" land = True weight = 9 skip_iav = True [DOLCE] source = "DATA/evspsbl/DOLCE/DOLCE.nc" weight = 15 land = True #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Sensible Heat] variable = "hfss" alternate_vars = "sh" weight = 2 mass_weighting = True [FLUXCOM] source = "DATA/hfss/FLUXCOM/sh.nc" weight = 15 skip_iav = True ########################################################################### [h1: Radiation and Energy Cycle] bgcolor = "#FFECE6" #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Albedo] variable = "albedo" weight = 1 ctype = "ConfAlbedo" [CERESed4.1] source = "DATA/albedo/CERESed4.1/albedo.nc" weight = 20 [GEWEX.SRB] source = "DATA/albedo/GEWEX.SRB/albedo_0.5x0.5.nc" weight = 20 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Upward SW Radiation] variable = "rsus" weight = 1 [FLUXNET2015] source = "DATA/rsus/FLUXNET2015/rsus.nc" weight = 12 [GEWEX.SRB] source = "DATA/rsus/GEWEX.SRB/rsus_0.5x0.5.nc" weight = 15 [WRMC.BSRN] source = "DATA/rsus/WRMC.BSRN/rsus.nc" weight = 12 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Net SW Radiation] variable = "rsns" derived = "rsds-rsus" weight = 1 [CERESed4.1] source = "DATA/rsns/CERESed4.1/rsns.nc" weight = 15 [FLUXNET2015] source = "DATA/rsns/FLUXNET2015/rsns.nc" weight = 12 [GEWEX.SRB] source = "DATA/rsns/GEWEX.SRB/rsns_0.5x0.5.nc" weight = 15 [WRMC.BSRN] source = "DATA/rsns/WRMC.BSRN/rsns.nc" weight = 12 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Upward LW Radiation] variable = "rlus" weight = 1 [FLUXNET2015] source = "DATA/rlus/FLUXNET2015/rlus.nc" weight = 12 [GEWEX.SRB] source = "DATA/rlus/GEWEX.SRB/rlus_0.5x0.5.nc" weight = 15 [WRMC.BSRN] source = "DATA/rlus/WRMC.BSRN/rlus.nc" weight = 12 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Net LW Radiation] variable = "rlns" derived = "rlds-rlus" weight = 1 [CERESed4.1] source = "DATA/rlns/CERESed4.1/rlns.nc" weight = 15 [FLUXNET2015] source = "DATA/rlns/FLUXNET2015/rlns.nc" weight = 12 [GEWEX.SRB] source = "DATA/rlns/GEWEX.SRB/rlns_0.5x0.5.nc" weight = 15 [WRMC.BSRN] source = "DATA/rlns/WRMC.BSRN/rlns.nc" weight = 12 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Net Radiation] variable = "rns" derived = "rlds-rlus+rsds-rsus" weight = 2 [CERESed4.1] source = "DATA/rns/CERESed4.1/rns.nc" weight = 15 [FLUXNET2015] source = "DATA/rns/FLUXNET2015/rns.nc" weight = 12 [GEWEX.SRB] source = "DATA/rns/GEWEX.SRB/rns_0.5x0.5.nc" weight = 15 [WRMC.BSRN] source = "DATA/rns/WRMC.BSRN/rns.nc" weight = 12 ########################################################################### [h1: Forcings] bgcolor = "#EDEDED" #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Air Temperature] variable = "tas" weight = 2 [FLUXNET2015] source = "DATA/tas/FLUXNET2015/tas.nc" weight = 9 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Diurnal Temperature Range] variable = "dtr" weight = 2 derived = "tasmax-tasmin" [CRU4.02] source = "DATA/dtr/CRU4.02/dtr.nc" weight = 25 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Precipitation] variable = "pr" cmap = "Blues" weight = 2 mass_weighting = True [FLUXNET2015] source = "DATA/pr/FLUXNET2015/pr.nc" land = True weight = 9 table_unit = "mm d-1" plot_unit = "mm d-1" [GPCCv2018] source = "DATA/pr/GPCCv2018/pr.nc" land = True weight = 20 table_unit = "mm d-1" plot_unit = "mm d-1" space_mean = True [GPCPv2.3] source = "DATA/pr/GPCPv2.3/pr.nc" land = True weight = 20 table_unit = "mm d-1" plot_unit = "mm d-1" space_mean = True #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Relative Humidity] variable = "rhums" alternate_vars = "hurs" cmap = "Blues" weight = 3 mass_weighting = True [CRU4.02] source = "DATA/rhums/CRU4.02/rhums.nc" weight = 10 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Downward SW Radiation] variable = "rsds" weight = 2 [FLUXNET2015] source = "DATA/rsds/FLUXNET2015/rsds.nc" weight = 12 [GEWEX.SRB] source = "DATA/rsds/GEWEX.SRB/rsds_0.5x0.5.nc" weight = 15 [WRMC.BSRN] source = "DATA/rsds/WRMC.BSRN/rsds.nc" weight = 12 #~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [h2: Surface Downward LW Radiation] variable = "rlds" weight = 1 [FLUXNET2015] source = "DATA/rlds/FLUXNET2015/rlds.nc" weight = 12 [GEWEX.SRB] source = "DATA/rlds/GEWEX.SRB/rlds_0.5x0.5.nc" weight = 15 [WRMC.BSRN] source = "DATA/rlds/WRMC.BSRN/rlds.nc" weight = 12
Running the ILAMB
Now that we have the configuration file set up, you can run the study using the ilamb-run script via the aforementioned
ilamb-run --config config.cfg --model_setup model_setup.txt --regions global
Taking advantage of multiprocessors
Because of the computational costs, you need to run ILAMB through a Portable Batch System (PBS) job on Gadi.
The following default PBS file, let’s call it ilamb_test.job, can help you to setup your own, while making sure to use the correct project (#PBS -P) to charge your computing cost to:
#!/bin/bash
#PBS -N ilamb_test
#PBS -l wd
#PBS -P your_compute_project_here
#PBS -q normalbw
#PBS -l walltime=0:20:00
#PBS -l ncpus=14
#PBS -l mem=63GB
#PBS -l jobfs=10GB
#PBS -l storage=gdata/ct11+gdata/hh5+gdata/xp65+gdata/fs38+gdata/oi10+gdata/zv30
# ILAMB is provided through projects xp65. We will use the latter here
module use /g/data/xp65/public/modules
module load conda/access-med
# Define the ILAMB Path, expecting it to be where you start this job from
export ILAMB_ROOT=./
export CARTOPY_DATA_DIR=/g/data/xp65/public/apps/cartopy-data
# Run ILAMB in parallel with the config.cfg configure file for the models defined in model_setup.txt
mpiexec -n 10 ilamb-run --config config.cfg --model_setup model_setup.txt --regions global
You should adjust this file to your own specifications (including the storage access to your models). Save the file in the $ILAMB_ROOT and submit its job to the queue from there via
qsub ilamb_test.job
Running this job will create a _build directory with the comparison results within $ILAMB_ROOT. You can adjust the place of this directory via a agrument --build_dir argument for ilamb-run.
View Result
Once you finish your ILAMB run, you will get your ILAMB result. The default path to the result is ./_built,unless you specified --build_dir before you run your experiment with ILAMB.
Use VSCode to Simplily Visualise
This is the recommended way to visualise result, you need to install extension Live Server in your VSCode, Type live server into the extensions search bar and select the Live Server extension published by Ritwick Dey marked with the red ellipse below. This extension allows us to preview html files from a browser on our computer, and will update automatically as the html file is updated in VS Code. We will use this extension to preview some of the ESMValTool recipe outputs that come in html format. Click install to add the extension.
Once you installed the extension, go to your result directory and right click index.html, choose Open with Live Server, then you will have your result opened in your browser.
In case you don’t use VSCode or Live Server doesn’t work for you, this is another way to view the result. You change your directory to the result directory, create a new local host by below command:
python3 -m http.server
Your ILAMB result can be viewed in the following address: localhost address:
http://0.0.0.0:8000/
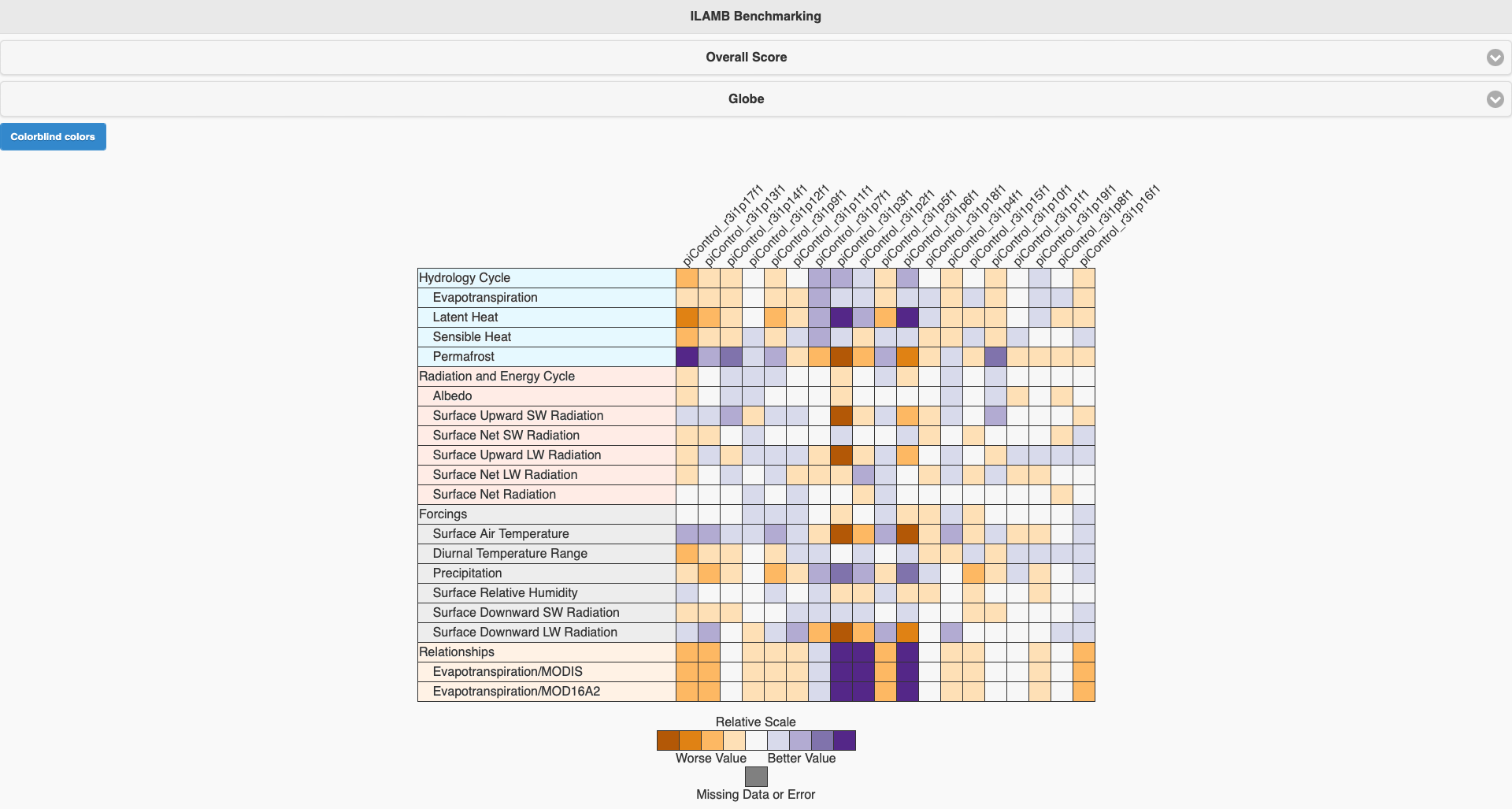
This is an example that we compared different ensemble members in piControl use the config.cfg we showed above. Click each raw, you can see the detail of conparison result with each observational data.
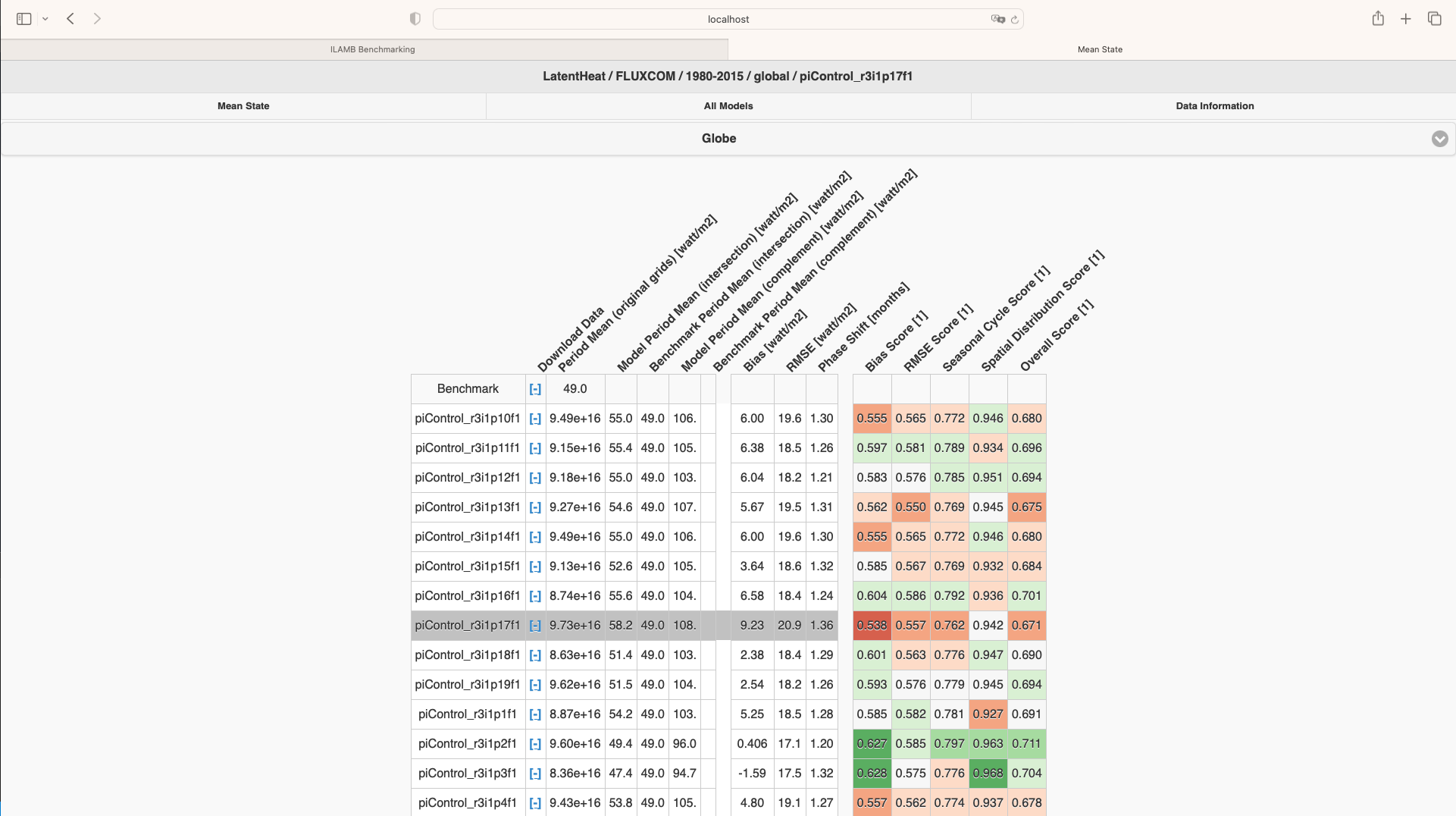
Click each raw of this matrix, you can view all the graphs of comparison results of this specific dataset.
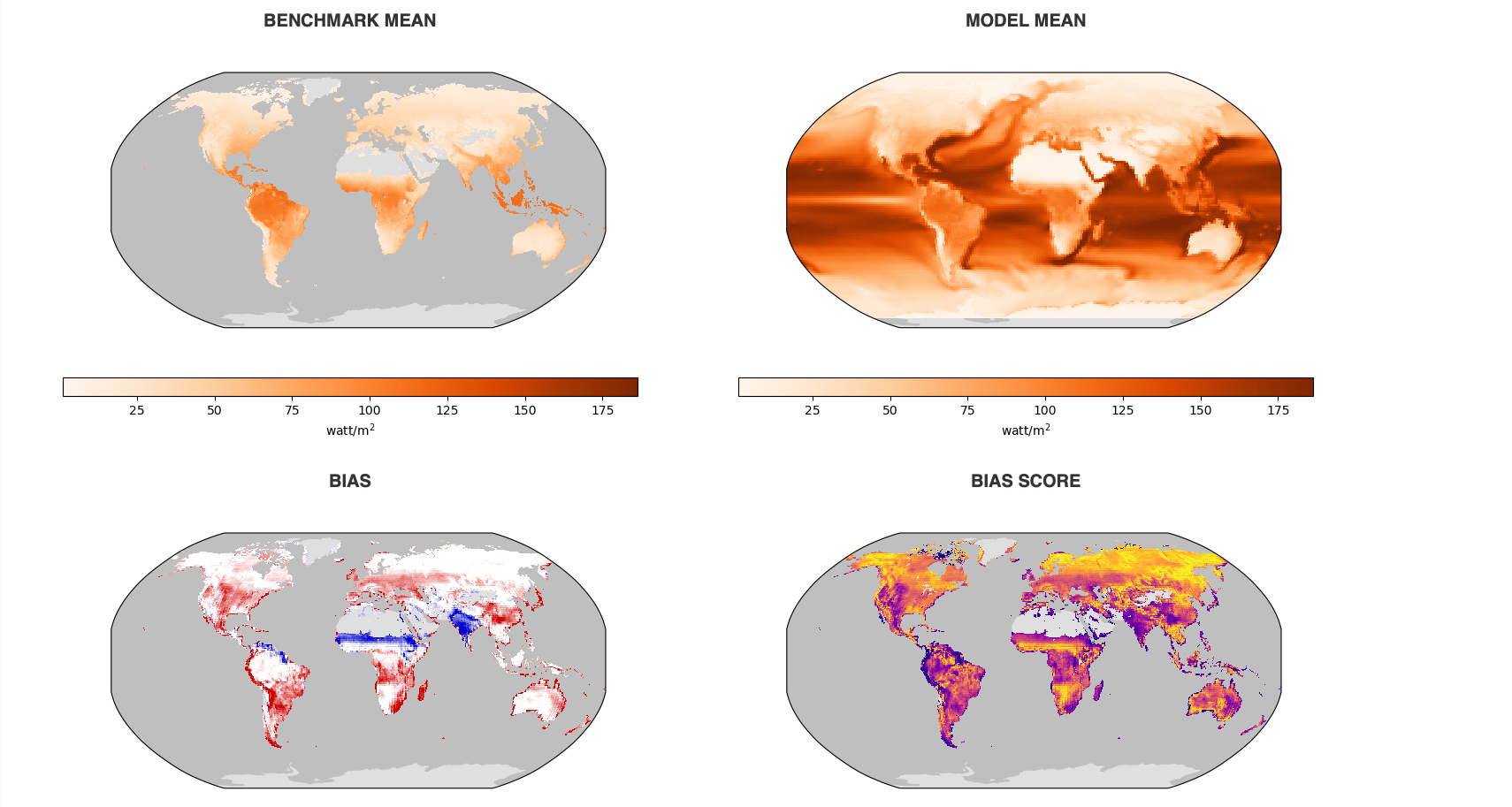
And also if you would like to view all graphs of one specific comparasion, you can click All Models, and choose which comparison you would liek to see(use Temporally integrated period mean rmse score as example), then you will get then all togather.
Key Points
The ACCESS-NRI ILAMB-Workflow facilitates the configuration of the ILAMB on NCI Gadi.
Users need to set up a run using a configuration file.
The
ilamb-tree-generatorallows to quickly build a data directory srtucture for the ILAMB.The ILAMB can take advantage of the multiple CPUs available on Gadi.
ILAMB support for RAW ACCESS-ESM outputs
Overview
Teaching: 15 min
Exercises: 15 min
Compatibility:Questions
What do we mean by CMORising?
How to use ilamb-tree-generator to CMORise Raw Access data
Objectives
Analyse raw (non-CMORised) ACCESS outputs with the ILAMB
In this episode we will introduce how to use ilamb-tree-generator as a CMORiser to help you use ILAMB to evaluate Access raw output. But before that, we will introduce what is ‘CMORise’ first.
What is CMORisation?
“CMORise” refers to the process of converting climate model output data into a standardized format that conforms to the Climate and Forecast (CF) metadata conventions. This process involves using the Climate Model Output Rewriter (CMOR) tool, which ensures that the data adheres to specific requirements for structure, metadata, and units, making it easier to compare and share across different climate models.
Use ilamb-tree-generator to CMORise Access raw output
Load the ILAMB-Workflow module
Theilamb-tree-generator is available in the ILAMB-Workflow module that can be loaded as follow:
module use /g/data/xp65/public/modules
module load ilamb-workflow
or
module use /g/data/xp65/public/modules
module load conda/access-med
Configuring Dataset Inputs for ilamb-tree-generator: CMIP and Non-CMIP Examples”
As mentioned earlier, the ilamb-tree-generator utilizes a .yml file for all input configurations. This format is consistent for different datasets. Below is an example configuration for both CMIP and non-CMIP datasets:
datasets:
- {mip: CMIP, institute: CSIRO, dataset: ACCESS-ESM1-5, project: CMIP6, exp: historical, ensemble: r1i1p1f1}
- {mip: non-CMIP, institute: CSIRO, dataset: ACCESS-ESM1-5, project: CMIP6, exp: HI-CN-05}
The first entry represents a CMIP dataset, which is the standard usage for ilamb-tree-generator. The second entry corresponds to an ACCESS raw output, which is a non-CMIP dataset. Although most parameters are similar, there are specific settings for non-CMIP datasets. Here are the details of each parameter:
mip: Set tonon-CMIPto activate the CMORiser for non-CMIP data.path: For users working with their own ACCESS raw data, specify the root directory here. If not provided, the tool will default to using data in thep73directory.
run ilamb-tree-generator
After setting up the config.yml file, run the ilamb-tree-generator. This will generate the CMORized data within the ILAMB-ROOT directory, making it accessible for ILAMB to read and use:
ilamb-tree-generator --datasets {your-config.yml-file} --ilamb_root $ILAMB_ROOT
Once it finish, you will get your CMORised data been stored by variable names in this format:
.
├── DATA
└── MODELS
└── ACCESS-ESM1-5
└── HI-CN-05
├── cSoil.nc
├── cVeg.nc
├── evspsbl.nc
├── gpp.nc
├── hfls.nc
├── hfss.nc
├── hurs.nc
├── lai.nc
├── nbp.nc
├── pr.nc
├── ra.nc
├── rh.nc
├── rlds.nc
├── rlus.nc
├── rsds.nc
├── rsus.nc
├── tasmax.nc
├── tasmin.nc
├── tas.nc
└── tsl.nc
Limitations
ilamb-tree-generatordoesn’t support all variable inACCESS-ESM1-5, only 19 variables which is required inilamb.cfg. Will try to add more variables in the next version.
Key Points
The ILAMB-Workflow only support RAW ACCESS data
Running the ILAMB-Workflow on RAW ACCESS data can take some time. Consider if it is appropriate for your work
Only a limited number of CMIP variables are supported