Running your first recipe
Overview
Teaching: 15 min
Exercises: 15 min
Compatibility: ESMValTool v2.11.0Questions
How to run a recipe?
What happens when I run a recipe?
Objectives
Run an existing ESMValTool recipe
Examine the log information
Navigate the output created by ESMValTool
Make small adjustments to an existing recipe
This episode describes how ESMValTool recipes work, how to run a recipe and how to explore the recipe output. By the end of this episode, you should be able to run your first recipe, look at the recipe output, and make small modifications.
Import module in GADI
You may want to open VS Code with a remote SSH connection to Gadi and use the VS Code terminal, then you can later view the recipe file. Refer to VS Code setup.
In a terminal with an SSH connection into Gadi, load the module to use ESMValTool on Gadi.
module use /g/data/xp65/public/modules
module load esmvaltool-workflow
Running an existing recipe
The recipe format has briefly been introduced in the Introduction episode. To see all the recipes that are shipped with ESMValTool, type
esmvaltool recipes list
We will start by running examples/recipe_python.yml. This is the command with ESMValTool installed.
esmvaltool run examples/recipe_python.yml
On Gadi, this can be done using the esmvaltool-workflow wrapper in the loaded module.
esmvaltool-workflow run examples/recipe_python.yml
or if you have the user configuration file in your current directory then
esmvaltool-workflow run --config_file ./config-user.yml examples/recipe_python.yml
You should see that Gadi has created a PBS job to run the recipe. You can check your
queue status with qstat.
[fc6164@gadi-login-01 fc6164]$ module load esmvaltool
Welcome to the ACCESS-NRI ESMValTool-Workflow
enter command `esmvaltool-workflow` for help
Loading esmvaltool/workflow_v1.2
Loading requirement: singularity conda/esmvaltool-0.4
[fc6164@gadi-login-01 fc6164]$ esmvaltool-workflow run recipe_python.yml
conda/esmvaltool-0.4
123732363.gadi-pbs
Running recipe: recipe_python.yml
[fc6164@gadi-login-01 fc6164]$ qstat
Job id Name User Time Use S Queue
--------------------- ---------------- ---------------- -------- - -----
123732363.gadi-pbs recipe_python fc6164 0 Q normal-exec
[fc6164@gadi-login-01 fc6164]$
If everything is okay, the final log message should be “Run was successful”. The exact output varies depending on your machine, this is an example of a successful log output below.
Example output
2024-05-15 07:04:08,041 UTC [134535] INFO ______________________________________________________________________ _____ ____ __ ____ __ _ _____ _ | ____/ ___|| \/ \ \ / /_ _| |_ _|__ ___ | | | _| \___ \| |\/| |\ \ / / _` | | | |/ _ \ / _ \| | | |___ ___) | | | | \ V / (_| | | | | (_) | (_) | | |_____|____/|_| |_| \_/ \__,_|_| |_|\___/ \___/|_| ______________________________________________________________________ ESMValTool - Earth System Model Evaluation Tool. http://www.esmvaltool.org CORE DEVELOPMENT TEAM AND CONTACTS: Birgit Hassler (Co-PI; DLR, Germany - birgit.hassler@dlr.de) Alistair Sellar (Co-PI; Met Office, UK - alistair.sellar@metoffice.gov.uk) Bouwe Andela (Netherlands eScience Center, The Netherlands - b.andela@esciencecenter.nl) Lee de Mora (PML, UK - ledm@pml.ac.uk) Niels Drost (Netherlands eScience Center, The Netherlands - n.drost@esciencecenter.nl) Veronika Eyring (DLR, Germany - veronika.eyring@dlr.de) Bettina Gier (UBremen, Germany - gier@uni-bremen.de) Remi Kazeroni (DLR, Germany - remi.kazeroni@dlr.de) Nikolay Koldunov (AWI, Germany - nikolay.koldunov@awi.de) Axel Lauer (DLR, Germany - axel.lauer@dlr.de) Saskia Loosveldt-Tomas (BSC, Spain - saskia.loosveldt@bsc.es) Ruth Lorenz (ETH Zurich, Switzerland - ruth.lorenz@env.ethz.ch) Benjamin Mueller (LMU, Germany - b.mueller@iggf.geo.uni-muenchen.de) Valeriu Predoi (URead, UK - valeriu.predoi@ncas.ac.uk) Mattia Righi (DLR, Germany - mattia.righi@dlr.de) Manuel Schlund (DLR, Germany - manuel.schlund@dlr.de) Breixo Solino Fernandez (DLR, Germany - breixo.solinofernandez@dlr.de) Javier Vegas-Regidor (BSC, Spain - javier.vegas@bsc.es) Klaus Zimmermann (SMHI, Sweden - klaus.zimmermann@smhi.se) For further help, please read the documentation at http://docs.esmvaltool.org. Have fun! 2024-05-15 07:04:08,044 UTC [134535] INFO Package versions 2024-05-15 07:04:08,044 UTC [134535] INFO ---------------- 2024-05-15 07:04:08,044 UTC [134535] INFO ESMValCore: 2.10.0 2024-05-15 07:04:08,044 UTC [134535] INFO ESMValTool: 2.10.0 2024-05-15 07:04:08,044 UTC [134535] INFO ---------------- 2024-05-15 07:04:08,044 UTC [134535] INFO Using config file /pfs/lustrep1/users/username/esmvaltool_tutorial/config-user.yml 2024-05-15 07:04:08,044 UTC [134535] INFO Writing program log files to: /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/main_log.txt /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/main_log_debug.txt 2024-05-15 07:04:08,503 UTC [134535] INFO Using default ESGF configuration, configuration file /users/username/.esmvaltool/esgf-pyclient.yml not present. 2024-05-15 07:04:08,504 UTC [134535] WARNING ESGF credentials missing, only data that is accessible without logging in will be available. See https://esgf.github.io/esgf-user-support/user_guide.html for instructions on how to create an account if you do not have one yet. Next, configure your system so esmvaltool can use your credentials. This can be done using the keyring package, or you can just enter them in /users/username/.esmvaltool/esgf-pyclient.yml. keyring ======= First install the keyring package (requires a supported backend, see https://pypi.org/project/keyring/): $ pip install keyring Next, set your username and password by running the commands: $ keyring set ESGF hostname $ keyring set ESGF username $ keyring set ESGF password To check that you entered your credentials correctly, run: $ keyring get ESGF hostname $ keyring get ESGF username $ keyring get ESGF password configuration file ================== You can store the hostname, username, and password or your OpenID account in a plain text in the file /users/username/.esmvaltool/esgf-pyclient.yml like this: logon: hostname: "your-hostname" username: "your-username" password: "your-password" or your can configure an interactive log in: logon: interactive: true Note that storing your password in plain text in the configuration file is less secure. On shared systems, make sure the permissions of the file are set so only you can read it, i.e. $ ls -l /users/username/.esmvaltool/esgf-pyclient.yml shows permissions -rw-------. 2024-05-15 07:04:09,067 UTC [134535] INFO Starting the Earth System Model Evaluation Tool at time: 2024-05-15 07:04:09 UTC 2024-05-15 07:04:09,068 UTC [134535] INFO ---------------------------------------------------------------------- 2024-05-15 07:04:09,068 UTC [134535] INFO RECIPE = /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/recipes/examples/recipe_python.yml 2024-05-15 07:04:09,068 UTC [134535] INFO RUNDIR = /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run 2024-05-15 07:04:09,069 UTC [134535] INFO WORKDIR = /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/work 2024-05-15 07:04:09,069 UTC [134535] INFO PREPROCDIR = /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/preproc 2024-05-15 07:04:09,069 UTC [134535] INFO PLOTDIR = /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/plots 2024-05-15 07:04:09,069 UTC [134535] INFO ---------------------------------------------------------------------- 2024-05-15 07:04:09,069 UTC [134535] INFO Running tasks using at most 256 processes 2024-05-15 07:04:09,069 UTC [134535] INFO If your system hangs during execution, it may not have enough memory for keeping this number of tasks in memory. 2024-05-15 07:04:09,070 UTC [134535] INFO If you experience memory problems, try reducing 'max_parallel_tasks' in your user configuration file. 2024-05-15 07:04:09,070 UTC [134535] WARNING Using the Dask basic scheduler. This may lead to slow computations and out-of-memory errors. Note that the basic scheduler may still be the best choice for preprocessor functions that are not lazy. In that case, you can safely ignore this warning. See https://docs.esmvaltool.org/projects/ESMValCore/en/latest/quickstart/configure.html#dask-distributed-configuration for more information. 2024-05-15 07:04:09,113 UTC [134535] WARNING 'default' rootpaths '/users/username/climate_data' set in config-user.yml do not exist 2024-05-15 07:04:10,648 UTC [134535] INFO Creating tasks from recipe 2024-05-15 07:04:10,648 UTC [134535] INFO Creating tasks for diagnostic map 2024-05-15 07:04:10,648 UTC [134535] INFO Creating diagnostic task map/script1 2024-05-15 07:04:10,649 UTC [134535] INFO Creating preprocessor task map/tas 2024-05-15 07:04:10,649 UTC [134535] INFO Creating preprocessor 'to_degrees_c' task for variable 'tas' 2024-05-15 07:04:11,066 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP6, BCC-ESM1, CMIP, historical, r1i1p1f1, gn, v20181214 2024-05-15 07:04:11,405 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP5, bcc-csm1-1, historical, r1i1p1, v1 2024-05-15 07:04:11,406 UTC [134535] INFO PreprocessingTask map/tas created. 2024-05-15 07:04:11,406 UTC [134535] INFO Creating tasks for diagnostic timeseries 2024-05-15 07:04:11,406 UTC [134535] INFO Creating diagnostic task timeseries/script1 2024-05-15 07:04:11,406 UTC [134535] INFO Creating preprocessor task timeseries/tas_amsterdam 2024-05-15 07:04:11,406 UTC [134535] INFO Creating preprocessor 'annual_mean_amsterdam' task for variable 'tas_amsterdam' 2024-05-15 07:04:11,428 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP6, BCC-ESM1, CMIP, historical, r1i1p1f1, gn, v20181214 2024-05-15 07:04:11,452 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP5, bcc-csm1-1, historical, r1i1p1, v1 2024-05-15 07:04:11,455 UTC [134535] INFO PreprocessingTask timeseries/tas_amsterdam created. 2024-05-15 07:04:11,455 UTC [134535] INFO Creating preprocessor task timeseries/tas_global 2024-05-15 07:04:11,455 UTC [134535] INFO Creating preprocessor 'annual_mean_global' task for variable 'tas_global' 2024-05-15 07:04:11,814 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP6, BCC-ESM1, CMIP, historical, r1i1p1f1, gn, v20181214, supplementaries: areacella, fx, 1pctCO2, v20190613 2024-05-15 07:04:12,184 UTC [134535] INFO Found input files for Dataset: tas, Amon, CMIP5, bcc-csm1-1, historical, r1i1p1, v1, supplementaries: areacella, fx, r0i0p0 2024-05-15 07:04:12,186 UTC [134535] INFO PreprocessingTask timeseries/tas_global created. 2024-05-15 07:04:12,187 UTC [134535] INFO These tasks will be executed: timeseries/script1, timeseries/tas_global, map/script1, map/tas, timeseries/tas_amsterdam 2024-05-15 07:04:12,204 UTC [134535] INFO Wrote recipe with version numbers and wildcards to: file:///users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/recipe_python_filled.yml 2024-05-15 07:04:12,204 UTC [134535] INFO Will download 129.2 MB Will download the following files: 50.85 KB ESGFFile:CMIP6/CMIP/BCC/BCC-ESM1/1pctCO2/r1i1p1f1/fx/areacella/gn/v20190613/areacella_fx_BCC-ESM1_1pctCO2_r1i1p1f1_gn.nc on hosts ['aims3.llnl.gov', 'cmip.bcc.cma.cn', 'esgf-data04.diasjp.net', 'esgf.nci.org.au', 'esgf3.dkrz.de'] 64.95 MB ESGFFile:CMIP6/CMIP/BCC/BCC-ESM1/historical/r1i1p1f1/Amon/tas/gn/v20181214/tas_Amon_BCC-ESM1_historical_r1i1p1f1_gn_185001-201412.nc on hosts ['aims3.llnl.gov', 'cmip.bcc.cma.cn', 'esgf-data04.diasjp.net', 'esgf.ceda.ac.uk', 'esgf.nci.org.au', 'esgf3.dkrz.de'] 44.4 KB ESGFFile:cmip5/output1/BCC/bcc-csm1-1/historical/fx/atmos/fx/r0i0p0/v1/areacella_fx_bcc-csm1-1_historical_r0i0p0.nc on hosts ['aims3.llnl.gov', 'esgf.ceda.ac.uk', 'esgf2.dkrz.de'] 64.15 MB ESGFFile:cmip5/output1/BCC/bcc-csm1-1/historical/mon/atmos/Amon/r1i1p1/v1/tas_Amon_bcc-csm1-1_historical_r1i1p1_185001-201212.nc on hosts ['aims3.llnl.gov', 'esgf.ceda.ac.uk', 'esgf2.dkrz.de'] Downloading 129.2 MB.. 2024-05-15 07:04:14,074 UTC [134535] INFO Downloaded /users/username/climate_data/cmip5/output1/BCC/bcc-csm1-1/historical/fx/atmos/fx/r0i0p0/v1/areacella_fx_bcc-csm1-1_historical_r0i0p0.nc (44.4 KB) in 1.84 seconds (24.09 KB/s) from aims3.llnl.gov 2024-05-15 07:04:14,109 UTC [134535] INFO Downloaded /users/username/climate_data/CMIP6/CMIP/BCC/BCC-ESM1/1pctCO2/r1i1p1f1/fx/areacella/gn/v20190613/areacella_fx_BCC-ESM1_1pctCO2_r1i1p1f1_gn.nc (50.85 KB) in 1.88 seconds (27 KB/s) from aims3.llnl.gov 2024-05-15 07:04:20,505 UTC [134535] INFO Downloaded /users/username/climate_data/CMIP6/CMIP/BCC/BCC-ESM1/historical/r1i1p1f1/Amon/tas/gn/v20181214/tas_Amon_BCC-ESM1_historical_r1i1p1f1_gn_185001-201412.nc (64.95 MB) in 8.27 seconds (7.85 MB/s) from aims3.llnl.gov 2024-05-15 07:04:25,862 UTC [134535] INFO Downloaded /users/username/climate_data/cmip5/output1/BCC/bcc-csm1-1/historical/mon/atmos/Amon/r1i1p1/v1/tas_Amon_bcc-csm1-1_historical_r1i1p1_185001-201212.nc (64.15 MB) in 13.63 seconds (4.71 MB/s) from aims3.llnl.gov 2024-05-15 07:04:25,870 UTC [134535] INFO Downloaded 129.2 MB in 13.67 seconds (9.45 MB/s) 2024-05-15 07:04:25,870 UTC [134535] INFO Successfully downloaded all requested files. 2024-05-15 07:04:25,871 UTC [134535] INFO Using the Dask basic scheduler. 2024-05-15 07:04:25,871 UTC [134535] INFO Running 5 tasks using 5 processes 2024-05-15 07:04:25,956 UTC [144507] INFO Starting task map/tas in process [144507] 2024-05-15 07:04:25,956 UTC [144522] INFO Starting task timeseries/tas_amsterdam in process [144522] 2024-05-15 07:04:25,957 UTC [144534] INFO Starting task timeseries/tas_global in process [144534] 2024-05-15 07:04:26,049 UTC [134535] INFO Progress: 3 tasks running, 2 tasks waiting for ancestors, 0/5 done 2024-05-15 07:04:26,457 UTC [144534] WARNING Long name changed from 'Grid-Cell Area for Atmospheric Variables' to 'Grid-Cell Area for Atmospheric Grid Variables' (for file /users/username/climate_data/CMIP6/CMIP/BCC/BCC-ESM1/1pctCO2/r1i1p1f1/fx/areacella/gn/v20190613/areacella_fx_BCC-ESM1_1pctCO2_r1i1p1f1_gn.nc) 2024-05-15 07:04:26,461 UTC [144507] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:26,856 UTC [144522] INFO Extracting data for Amsterdam, Noord-Holland, Nederland (52.3730796 °N, 4.8924534 °E) 2024-05-15 07:04:27,081 UTC [144507] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:27,085 UTC [144534] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:40,666 UTC [144507] INFO Successfully completed task map/tas (priority 1) in 0:00:14.709864 2024-05-15 07:04:40,805 UTC [134535] INFO Progress: 2 tasks running, 2 tasks waiting for ancestors, 1/5 done 2024-05-15 07:04:40,813 UTC [144547] INFO Starting task map/script1 in process [144547] 2024-05-15 07:04:40,821 UTC [144547] INFO Running command ['/LUMI_TYKKY_D1Npoag/miniconda/envs/env1/bin/python', '/LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/diag_scripts/examples/diagnostic.py', '/users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/map/script1/settings.yml'] 2024-05-15 07:04:40,822 UTC [144547] INFO Writing output to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/work/map/script1 2024-05-15 07:04:40,822 UTC [144547] INFO Writing plots to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/plots/map/script1 2024-05-15 07:04:40,822 UTC [144547] INFO Writing log to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/map/script1/log.txt 2024-05-15 07:04:40,822 UTC [144547] INFO To re-run this diagnostic script, run: cd /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/map/script1; MPLBACKEND="Agg" /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/bin/python /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/diag_scripts/examples/diagnostic.py /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/map/script1/settings.yml 2024-05-15 07:04:40,906 UTC [134535] INFO Progress: 3 tasks running, 1 tasks waiting for ancestors, 1/5 done 2024-05-15 07:04:47,225 UTC [144522] INFO Extracting data for Amsterdam, Noord-Holland, Nederland (52.3730796 °N, 4.8924534 °E) 2024-05-15 07:04:47,308 UTC [144534] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:47,697 UTC [144534] INFO Successfully completed task timeseries/tas_global (priority 4) in 0:00:21.738941 2024-05-15 07:04:47,845 UTC [134535] INFO Progress: 2 tasks running, 1 tasks waiting for ancestors, 2/5 done 2024-05-15 07:04:48,053 UTC [144522] INFO Generated PreprocessorFile: /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/preproc/timeseries/tas_amsterdam/MultiModelMean_historical_Amon_tas_1850-2000.nc 2024-05-15 07:04:48,058 UTC [144522] WARNING /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/iris/fileformats/netcdf/saver.py:2670: IrisDeprecation: Saving to netcdf with legacy-style attribute handling for backwards compatibility. This mode is deprecated since Iris 3.8, and will eventually be removed. Please consider enabling the new split-attributes handling mode, by setting 'iris.FUTURE.save_split_attrs = True'. warn_deprecated(message) 2024-05-15 07:04:48,228 UTC [144522] INFO Successfully completed task timeseries/tas_amsterdam (priority 3) in 0:00:22.271045 2024-05-15 07:04:48,346 UTC [134535] INFO Progress: 1 tasks running, 1 tasks waiting for ancestors, 3/5 done 2024-05-15 07:04:48,358 UTC [144558] INFO Starting task timeseries/script1 in process [144558] 2024-05-15 07:04:48,364 UTC [144558] INFO Running command ['/LUMI_TYKKY_D1Npoag/miniconda/envs/env1/bin/python', '/LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/diag_scripts/examples/diagnostic.py', '/users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/timeseries/script1/settings.yml'] 2024-05-15 07:04:48,365 UTC [144558] INFO Writing output to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/work/timeseries/script1 2024-05-15 07:04:48,365 UTC [144558] INFO Writing plots to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/plots/timeseries/script1 2024-05-15 07:04:48,365 UTC [144558] INFO Writing log to /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/timeseries/script1/log.txt 2024-05-15 07:04:48,365 UTC [144558] INFO To re-run this diagnostic script, run: cd /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/timeseries/script1; MPLBACKEND="Agg" /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/bin/python /LUMI_TYKKY_D1Npoag/miniconda/envs/env1/lib/python3.11/site-packages/esmvaltool/diag_scripts/examples/diagnostic.py /users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/timeseries/script1/settings.yml 2024-05-15 07:04:48,447 UTC [134535] INFO Progress: 2 tasks running, 0 tasks waiting for ancestors, 3/5 done 2024-05-15 07:04:54,019 UTC [144547] INFO Maximum memory used (estimate): 0.4 GB 2024-05-15 07:04:54,021 UTC [144547] INFO Sampled every second. It may be inaccurate if short but high spikes in memory consumption occur. 2024-05-15 07:04:55,174 UTC [144547] INFO Successfully completed task map/script1 (priority 0) in 0:00:14.360271 2024-05-15 07:04:55,366 UTC [144558] INFO Maximum memory used (estimate): 0.4 GB 2024-05-15 07:04:55,368 UTC [144558] INFO Sampled every second. It may be inaccurate if short but high spikes in memory consumption occur. 2024-05-15 07:04:55,566 UTC [134535] INFO Progress: 1 tasks running, 0 tasks waiting for ancestors, 4/5 done 2024-05-15 07:04:56,958 UTC [144558] INFO Successfully completed task timeseries/script1 (priority 2) in 0:00:08.599797 2024-05-15 07:04:57,072 UTC [134535] INFO Progress: 0 tasks running, 0 tasks waiting for ancestors, 5/5 done 2024-05-15 07:04:57,072 UTC [134535] INFO Successfully completed all tasks. 2024-05-15 07:04:57,134 UTC [134535] INFO Wrote recipe with version numbers and wildcards to: file:///users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/run/recipe_python_filled.yml 2024-05-15 07:04:57,399 UTC [134535] INFO Wrote recipe output to: file:///users/username/esmvaltool_tutorial/esmvaltool_output/recipe_python_20240515_070408/index.html 2024-05-15 07:04:57,399 UTC [134535] INFO Ending the Earth System Model Evaluation Tool at time: 2024-05-15 07:04:57 UTC 2024-05-15 07:04:57,400 UTC [134535] INFO Time for running the recipe was: 0:00:48.332409 2024-05-15 07:04:57,756 UTC [134535] INFO Maximum memory used (estimate): 2.5 GB 2024-05-15 07:04:57,757 UTC [134535] INFO Sampled every second. It may be inaccurate if short but high spikes in memory consumption occur. 2024-05-15 07:04:57,759 UTC [134535] INFO Removing `preproc` directory containing preprocessed data 2024-05-15 07:04:57,759 UTC [134535] INFO If this data is further needed, then set `remove_preproc_dir` to `false` in your user configuration file 2024-05-15 07:04:57,782 UTC [134535] INFO Run was successful
On Gadi with esmvaltool-workflow you will see the wrapper has run esmvaltool in a
PBS job for you, when complete you can find the output in
/scratch/nf33/$USER/esmvaltool_outputs/. In the run folder, the main_log would
be the terminal output of the command. This recipe won’t complete as it needs internet
connection to search for the location.
We will modify this recipe later so that it completes, for now you will likely see the below in your log file.
Error output
ERROR [2488385] Program terminated abnormally, see stack trace below for more information: multiprocessing.pool.RemoteTraceback: """ Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connection.py", line 196, in _new_conn sock = connection.create_connection( ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/util/connection.py", line 85, in create_connection raise err File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/util/connection.py", line 73, in create_connection sock.connect(sa) OSError: [Errno 101] Network is unreachable The above exception was the direct cause of the following exception: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 789, in urlopen response = self._make_request( ^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 490, in _make_request raise new_e File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 466, in _make_request self._validate_conn(conn) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 1095, in _validate_conn conn.connect() File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connection.py", line 615, in connect self.sock = sock = self._new_conn() ^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connection.py", line 211, in _new_conn raise NewConnectionError( urllib3.exceptions.NewConnectionError: <urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable The above exception was the direct cause of the following exception: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/adapters.py", line 667, in send resp = conn.urlopen( ^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 873, in urlopen return self.urlopen( ^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 873, in urlopen return self.urlopen( ^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/connectionpool.py", line 843, in urlopen retries = retries.increment( ^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/urllib3/util/retry.py", line 519, in increment raise MaxRetryError(_pool, url, reason) from reason # type: ignore[arg-type] ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ urllib3.exceptions.MaxRetryError: HTTPSConnectionPool(host='nominatim.openstreetmap.org', port=443): Max retries exceeded with url: /search?q=Amsterdam&format=json&limit=1 (Caused by NewConnectionError('<urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable')) During handling of the above exception, another exception occurred: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/adapters.py", line 482, in _request resp = self.session.get(url, timeout=timeout, headers=headers) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/sessions.py", line 602, in get return self.request("GET", url, **kwargs) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/sessions.py", line 589, in request resp = self.send(prep, **send_kwargs) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/sessions.py", line 703, in send r = adapter.send(request, **kwargs) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/requests/adapters.py", line 700, in send raise ConnectionError(e, request=request) requests.exceptions.ConnectionError: HTTPSConnectionPool(host='nominatim.openstreetmap.org', port=443): Max retries exceeded with url: /search?q=Amsterdam&format=json&limit=1 (Caused by NewConnectionError('<urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable')) During handling of the above exception, another exception occurred: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/multiprocessing/pool.py", line 125, in worker result = (True, func(*args, **kwds)) ^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 816, in _run_task output_files = task.run() ^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 264, in run self.output_files = self._run(input_files) ^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/__init__.py", line 684, in _run product.apply(step, self.debug) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/__init__.py", line 492, in apply self.cubes = preprocess(self.cubes, step, ^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/__init__.py", line 401, in preprocess result.append(_run_preproc_function(function, item, settings, ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/__init__.py", line 346, in _run_preproc_function return function(items, **kwargs) ^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/preprocessor/_regrid.py", line 403, in extract_location geolocation = geolocator.geocode(location) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/geocoders/nominatim.py", line 297, in geocode return self._call_geocoder(url, callback, timeout=timeout) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/geocoders/base.py", line 368, in _call_geocoder result = self.adapter.get_json(url, timeout=timeout, headers=req_headers) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/adapters.py", line 472, in get_json resp = self._request(url, timeout=timeout, headers=headers) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/geopy/adapters.py", line 494, in _request raise GeocoderUnavailable(message) geopy.exc.GeocoderUnavailable: HTTPSConnectionPool(host='nominatim.openstreetmap.org', port=443): Max retries exceeded with url: /search?q=Amsterdam&format=json&limit=1 (Caused by NewConnectionError('<urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable')) """ The above exception was the direct cause of the following exception: Traceback (most recent call last): File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_main.py", line 533, in run fire.Fire(ESMValTool()) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/fire/core.py", line 143, in Fire component_trace = _Fire(component, args, parsed_flag_args, context, name) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/fire/core.py", line 477, in _Fire component, remaining_args = _CallAndUpdateTrace( ^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/fire/core.py", line 693, in _CallAndUpdateTrace component = fn(*varargs, **kwargs) ^^^^^^^^^^^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_main.py", line 413, in run self._run(recipe, session) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_main.py", line 455, in _run process_recipe(recipe_file=recipe, session=session) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_main.py", line 130, in process_recipe recipe.run() File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_recipe/recipe.py", line 1095, in run self.tasks.run(max_parallel_tasks=self.session['max_parallel_tasks']) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 738, in run self._run_parallel(address, max_parallel_tasks) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 782, in _run_parallel _copy_results(task, running[task]) File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/site-packages/esmvalcore/_task.py", line 805, in _copy_results task.output_files, task.products = future.get() ^^^^^^^^^^^^ File "/g/data/xp65/public/apps/med_conda/envs/esmvaltool-0.4/lib/python3.11/multiprocessing/pool.py", line 774, in get raise self._value geopy.exc.GeocoderUnavailable: HTTPSConnectionPool(host='nominatim.openstreetmap.org', port=443): Max retries exceeded with url: /search?q=Amsterdam&format=json&limit=1 (Caused by NewConnectionError('<urllib3.connection.HTTPSConnection object at 0x14fafc352e10>: Failed to establish a new connection: [Errno 101] Network is unreachable')) INFO [2488385] If you have a question or need help, please start a new discussion on https://github.com/ESMValGroup/ESMValTool/discussions If you suspect this is a bug, please open an issue on https://github.com/ESMValGroup/ESMValTool/issues To make it easier to find out what the problem is, please consider attaching the files run/recipe_*.yml and run/main_log_debug.txt from the output directory.
Pro tip: ESMValTool search paths
You might wonder how ESMValTool was able find the recipe file, even though it’s not in your working directory. All the recipe paths printed from
esmvaltool recipes listare relative to ESMValTool’s installation location. This is where ESMValTool will look if it cannot find the file by following the path from your working directory.
Investigating the log messages
Let’s dissect what’s happening here.
Output files and directories
After the banner and general information, the output starts with some important locations.
- Did ESMValTool use the right config file?
- What is the path to the example recipe?
- What is the main output folder generated by ESMValTool?
- Can you guess what the different output directories are for?
- ESMValTool creates two log files. What is the difference?
Answers
- The config file should be the one we edited in the previous episode, something like
/home/<username>/.esmvaltool/config-user.ymlor~/esmvaltool_tutorial/config-user.yml.- ESMValTool found the recipe in its installation directory, something like
/home/users/username/mambaforge/envs/esmvaltool/bin/esmvaltool/recipes/examples/or if you are using a pre-installed module on a server, something like/apps/jasmin/community/esmvaltool/ESMValTool_<version> /esmvaltool/recipes/examples/recipe_python.yml, where<version>is the latest release.- ESMValTool creates a time-stamped output directory for every run. In this case, it should be something like
recipe_python_YYYYMMDD_HHMMSS. This folder is made inside the output directory specified in the previous episode:~/esmvaltool_tutorial/esmvaltool_output.- There should be four output folders:
plots/: this is where output figures are stored.preproc/: this is where pre-processed data are stored.run/: this is where esmvaltool stores general information about the run, such as log messages and a copy of the recipe file.work/: this is where output files (not figures) are stored.- The log files are:
main_log.txtis a copy of the command-line outputmain_log_debug.txtcontains more detailed information that may be useful for debugging.
Debugging: No ‘preproc’ directory?
If you’re missing the preproc directory, then your
config-user.ymlfile has the valueremove_preproc_dirset totrue(this is used to save disk space). Please set this value tofalseand run the recipe again.
After the output locations, there are two main sections that can be distinguished in the log messages:
- Creating tasks
- Executing tasks
Analyse the tasks
List all the tasks that ESMValTool is executing for this recipe. Can you guess what this recipe does?
Answer
Just after all the ‘creating tasks’ and before ‘executing tasks’, we find the following line in the output:
[134535] INFO These tasks will be executed: map/tas, timeseries/tas_global, timeseries/script1, map/script1, timeseries/tas_amsterdamSo there are three tasks related to timeseries: global temperature, Amsterdam temperature, and a script (tas: near-surface air temperature). And then there are two tasks related to a map: something with temperature, and again a script.
Examining the recipe file
To get more insight into what is happening, we will have a look at the recipe
file itself. Use the following command to copy the recipe to your working
directory (eg. in \scratch\nf33\$USERNAME\)
esmvaltool recipes get examples/recipe_python.yml
Now you should see the recipe file in your working directory (type ls to
verify). Use VS Code to open this file, you should be able to open from your
explorer panel:
For reference, you can also view the recipe by unfolding the box below.
recipe_python.yml
# ESMValTool # recipe_python.yml # # See https://docs.esmvaltool.org/en/latest/recipes/recipe_examples.html # for a description of this recipe. # # See https://docs.esmvaltool.org/projects/esmvalcore/en/latest/recipe/overview.html # for a description of the recipe format. --- documentation: description: | Example recipe that plots a map and timeseries of temperature. title: Recipe that runs an example diagnostic written in Python. authors: - andela_bouwe - righi_mattia maintainer: - schlund_manuel references: - acknow_project projects: - esmval - c3s-magic datasets: - {dataset: BCC-ESM1, project: CMIP6, exp: historical, ensemble: r1i1p1f1, grid: gn} - {dataset: bcc-csm1-1, project: CMIP5, exp: historical, ensemble: r1i1p1} preprocessors: # See https://docs.esmvaltool.org/projects/esmvalcore/en/latest/recipe/preprocessor.html # for a description of the preprocessor functions. to_degrees_c: convert_units: units: degrees_C annual_mean_amsterdam: extract_location: location: Amsterdam scheme: linear annual_statistics: operator: mean multi_model_statistics: statistics: - mean span: overlap convert_units: units: degrees_C annual_mean_global: area_statistics: operator: mean annual_statistics: operator: mean convert_units: units: degrees_C diagnostics: map: description: Global map of temperature in January 2000. themes: - phys realms: - atmos variables: tas: mip: Amon preprocessor: to_degrees_c timerange: 2000/P1M caption: | Global map of {long_name} in January 2000 according to {dataset}. scripts: script1: script: examples/diagnostic.py quickplot: plot_type: pcolormesh cmap: Reds timeseries: description: Annual mean temperature in Amsterdam and global mean since 1850. themes: - phys realms: - atmos variables: tas_amsterdam: short_name: tas mip: Amon preprocessor: annual_mean_amsterdam timerange: 1850/2000 caption: Annual mean {long_name} in Amsterdam according to {dataset}. tas_global: short_name: tas mip: Amon preprocessor: annual_mean_global timerange: 1850/2000 caption: Annual global mean {long_name} according to {dataset}. scripts: script1: script: examples/diagnostic.py quickplot: plot_type: plot
Do you recognize the basic recipe structure that was introduced in episode 1?
- Documentation with relevant (citation) information
- Datasets that should be analysed
- Preprocessors groups of common preprocessing steps
- Diagnostics scripts performing more specific evaluation steps
Analyse the recipe
Try to answer the following questions:
- Who wrote this recipe?
- Who should be approached if there is a problem with this recipe?
- How many datasets are analyzed?
- What does the preprocessor called
annual_mean_globaldo?- Which script is applied for the diagnostic called
map?- Can you link specific lines in the recipe to the tasks that we saw before?
- How is the location of the city specified?
- How is the temporal range of the data specified?
Answers
- The example recipe is written by Bouwe Andela and Mattia Righi.
- Manuel Schlund is listed as the maintainer of this recipe.
- Two datasets are analysed:
- CMIP6 data from the model BCC-ESM1
- CMIP5 data from the model bcc-csm1-1
- The preprocessor
annual_mean_globalcomputes an area mean as well as annual means- The diagnostic called
mapexecutes a script referred to asscript1. This is a python script namedexamples/diagnostic.py- There are two diagnostics:
mapandtimeseries. Under the diagnosticmapwe find two tasks:
- a preprocessor task called
tas, applying the preprocessor calledto_degrees_cto the variabletas.- a diagnostic task called
script1, applying the scriptexamples/diagnostic.pyto the preprocessed data (map/tas).Under the diagnostic
timeserieswe find three tasks:
- a preprocessor task called
tas_amsterdam, applying the preprocessor calledannual_mean_amsterdamto the variabletas.- a preprocessor task called
tas_global, applying the preprocessor calledannual_mean_globalto the variabletas.- a diagnostic task called
script1, applying the scriptexamples/diagnostic.pyto the preprocessed data (timeseries/tas_globalandtimeseries/tas_amsterdam).- The
extract_locationpreprocessor is used to get data for a specific location here. ESMValTool interpolates to the location based on the chosen scheme. Can you tell the scheme used here? For more ways to extract areas, see the Area operations page.- The
timerangetag is used to extract data from a specific time period here. The start time is01/01/2000and the span of time to calculate means is1 Monthgiven byP1M. For more options on how to specify time ranges, see the timerange documentation.
Pro tip: short names and variable groups
The preprocessor tasks in ESMValTool are called ‘variable groups’. For the diagnostic
timeseries, we have two variable groups:tas_amsterdamandtas_global. Both of them operate on the variabletas(as indicated by theshort_name), but they apply different preprocessors. For the diagnosticmapthe variable group itself is namedtas, and you’ll notice that we do not explicitly provide theshort_name. This is a shorthand built into ESMValTool.
Output files
Have another look at the output directory created by the ESMValTool run.
Which files/folders are created by each task?
Answer
- map/tas: creates
/preproc/map/tas, which contains preprocessed data for each of the input datasets, a file calledmetadata.ymldescribing the contents of these datasets and provenance information in the form of.xmlfiles.- timeseries/tas_global: creates
/preproc/timeseries/tas_global, which contains preprocessed data for each of the input datasets, ametadata.ymlfile and provenance information in the form of.xmlfiles.- timeseries/tas_amsterdam: creates
/preproc/timeseries/tas_amsterdam, which contains preprocessed data for each of the input datasets, plus a combinedMultiModelMean, ametadata.ymlfile and provenance files.- map/script1: creates
/run/map/script1with general information and a log of the diagnostic script run. It also creates/plots/map/script1/and/work/map/script1, which contain output figures and output datasets, respectively. For each output file, there is also corresponding provenance information in the form of.xml,.bibtexand.txtfiles.- timeseries/script1: creates
/run/timeseries/script1with general information and a log of the diagnostic script run. It also creates/plots/timeseries/script1and/work/timeseries/script1, which contain output figures and output datasets, respectively. For each output file, there is also corresponding provenance information in the form of.xml,.bibtexand.txtfiles.
Pro tip: diagnostic logs
When you run ESMValTool, any log messages from the diagnostic script are not printed on the terminal. But they are written to the
log.txtfiles in the folder/run/<diag_name>/log.txt.ESMValTool does print a command that can be used to re-run a diagnostic script. When you use this the output will be printed to the command line.
Modifying the example recipe
Let’s make a small modification to the example recipe. Notice that now that you have copied and edited the recipe, you can use in your working directory:
esmvaltool-workflow run recipe_python.yml
to refer to your local file rather than the default version shipped with ESMValTool.
Change your location
Modify and run the recipe to analyse the temperature for your another location. Change the
extract_locationprerpocessor to one that doesn’t require internet connectionSolution
In principle, you only have to replace the
extract_locationwithextract_pointpreprocessor function and use latitude and longitude to define location. in the preprocessor calledannual_mean_amsterdam. However, it is good practice to also replace all instances ofamsterdamwith the correct name of your location. Otherwise the log messages and output will be confusing. You are free to modify the names of preprocessors or diagnostics.In the
difffile below you will see the changes we have made to the file. The top 2 lines are the filenames and the lines like@@ -39,9 +39,9 @@represent the line numbers in the original and modified file, respectively. For more info on this format, see here.--- recipe_python.yml +++ recipe_python_sydney.yml @@ -39,10 +39,9 @@ preprocessors: convert_units: units: degrees_C - annual_mean_amsterdam: - extract_location: - location: Amsterdam + annual_mean_sydney: + extract_point: + latitude: -34 + longitude: 151 scheme: linear annual_statistics: operator: mean @@ -84,18 +83,18 @@ diagnostics: themes: - phys realms: - atmos variables: - tas_amsterdam: + tas_sydney: short_name: tas mip: Amon - preprocessor: annual_mean_amsterdam + preprocessor: annual_mean_sydney timerange: 1850/2000 - caption: Annual mean {long_name} in Amsterdam according to {dataset}. + caption: Annual mean {long_name} in Sydney according to {dataset}. tas_global: short_name: tas mip: Amon
View the output
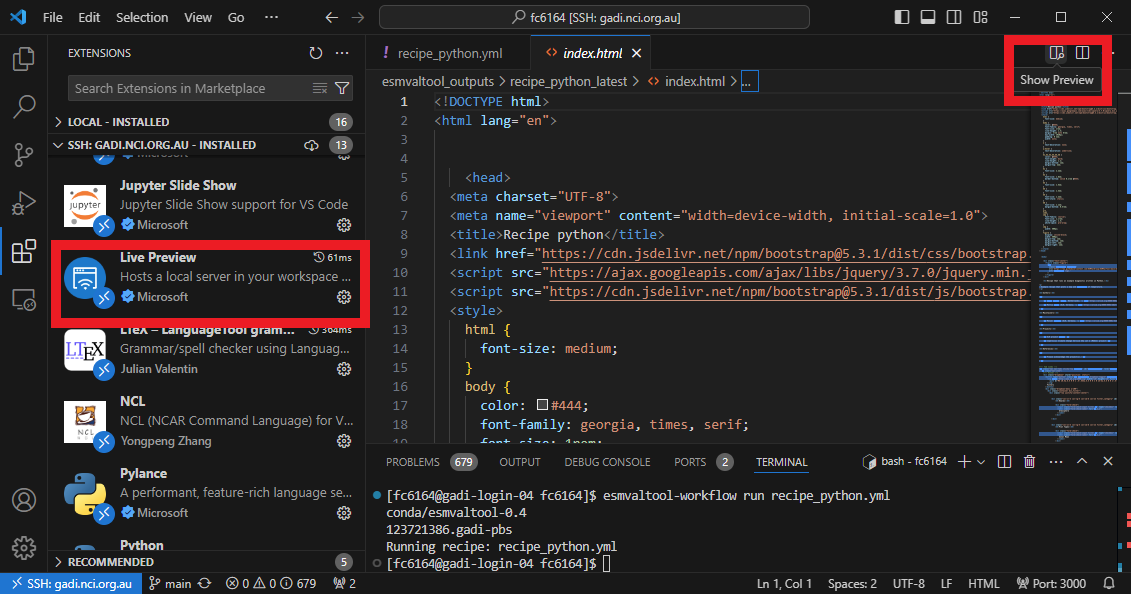
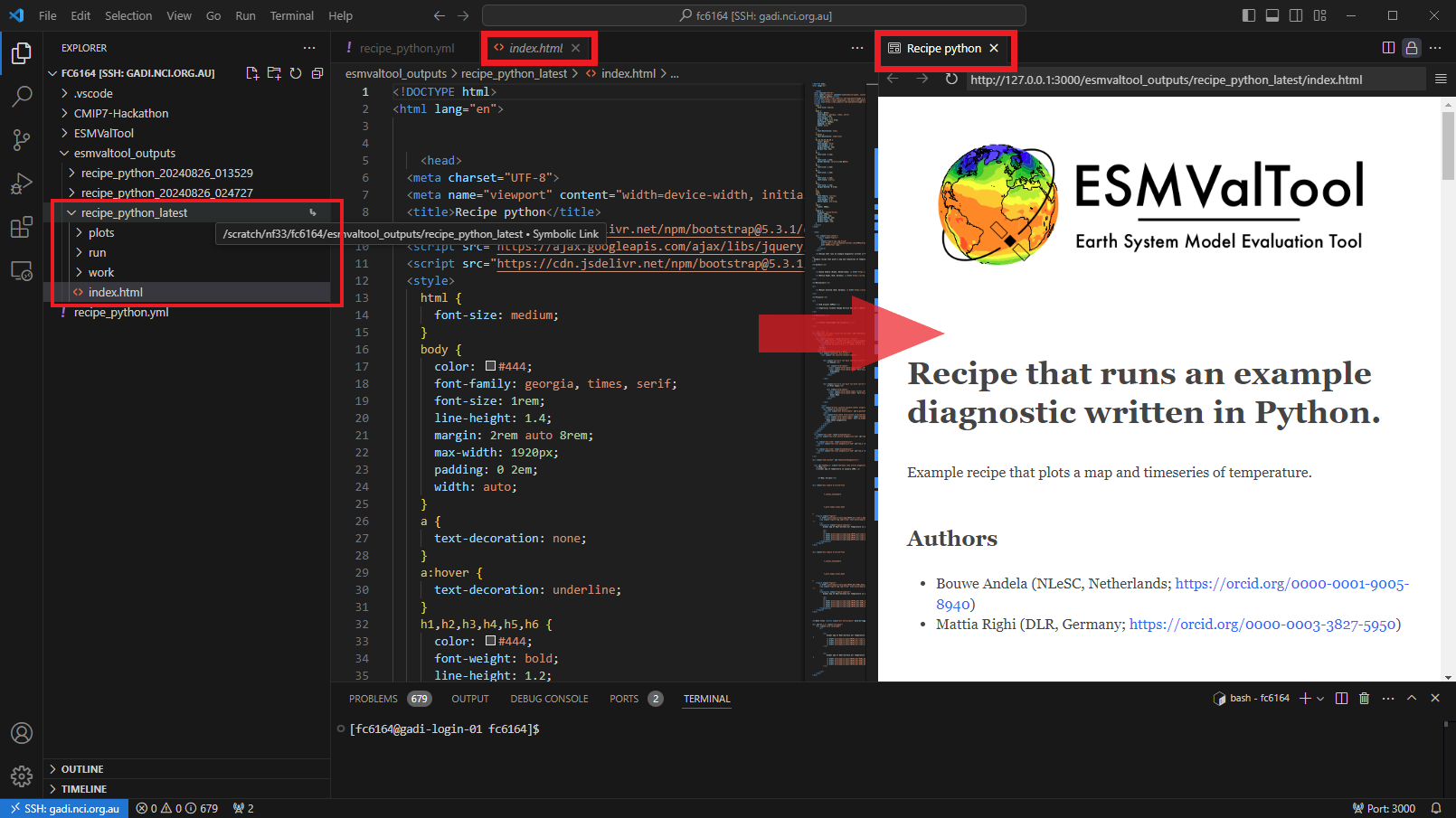
Now that the recipe runs we can look at the output. We recommend using VS Code with the “Live Preview” extension to view the html that is generated. When you open the html file, you will see the preview button appear in the top right.
Preview
You can see the output folder in explorer with the index.html file with a successful run. When you click on the preview button, the preview will appear to the right. You can also drag this across as a tab to use more of your screen to view.
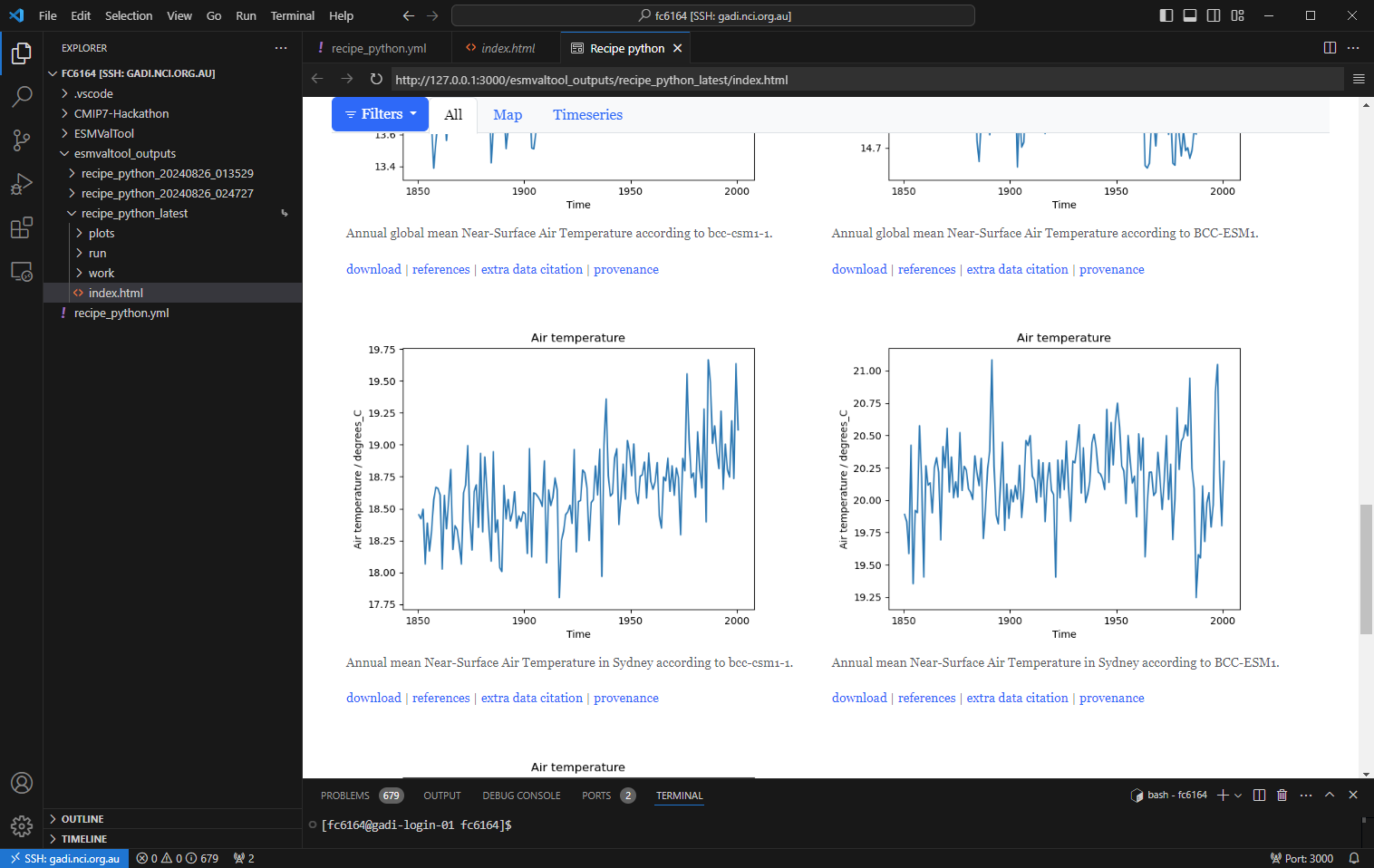
HTML output
Key Points
ESMValTool recipes work ‘out of the box’ (if input data is available)
There are strong links between the recipe, log file, and output folders
Recipes can easily be modified to re-use existing code for your own use case